The KLF4/Galectin-3 cascade is a key determinant of tubular cell death and acute kidney injury
- PMID: 41079936
- PMCID: PMC12509911
- DOI: 10.7150/ijbs.110790
The KLF4/Galectin-3 cascade is a key determinant of tubular cell death and acute kidney injury
Abstract
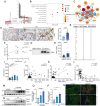
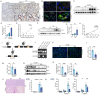
Clinically, acute kidney injury (AKI) stems from a diverse array of causes including ischemia, exposure to nephrotoxic agents, or sepsis. Renal tubular cells are particularly vulnerable and often sustain the most significant damage during AKI. This raises the question of whether there exists a common pathophysiological mechanism or pathway in renal tubular cells that underlies the development of AKI. We observed that tubular Galectin-3 is significantly up-regulated in four AKI mouse models and its tissue expression shows a positive correlation with tubular injury in human kidneys affected by AKI. The urinary Galectin-3 levels were markedly elevated in a cohort of patients with AKI and these levels correlated with the severity of kidney dysfunction. Based on predictions from bioinformatic analysis and JASPAR database, ChIP-PCR and luciferase-reporter assays demonstrated the direct binding of the transcription factor KLF4 to a specific sequence in the Galectin-3 gene promoter. Furthermore, mice with proximal tubular-specific deletion of KLF4 exhibited reduced kidney injury and inflammation, along with lower Galectin-3 expression in both cisplatin and ischemia-reperfusion-induced AKI. Targeting the KLF4/Galectin-3 axis with Kenpaullone and GB1107 confirmed protective effects against cisplatin-induced cell death and acute kidney injury, respectively. Our study highlights the KLF4/Galectin-3 pathway as a key mediator in the pathogenesis of AKI. Disrupting this signaling pathway may provide a promising therapeutic approach for the treatment of AKI.
Keywords: Galectin-3; KLF4; acute kidney injury; inflammation.; tubular cell death.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

