IFNγ regulates MR1 transcription and antigen presentation
- PMID: 41080531
- PMCID: PMC12510863
- DOI: 10.3389/fimmu.2025.1624767
IFNγ regulates MR1 transcription and antigen presentation
Abstract
Introduction: Antigen presentation molecules play key roles in T cell immunity. Multiple complementary pathways are known to regulate classical MHC-I molecules at transcriptional, translational, and post-translational levels. Intracellular trafficking mechanisms dictating post-transcriptional regulation of MR1, the MHC-I-like molecule which restricts MAIT cells, have been an area of focus; however, little is known about MR1 transcriptional regulation. We demonstrate that interferons regulate MR1 transcription.
Methods: Primary human airway epithelial cells (AEC) were treated with recombinant interferons or co-cultured with MAIT cell clones and antigen sources. MR1 expression was analyzed by RT-qPCR and flow cytometry. MAIT cell activity was quantified by ELISPOT.
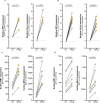
Results: Treatment of AECs with IFNβ or IFNγ variably increased MR1 transcripts, while only IFNγ significantly increased surface MR1 expression and enhanced antigen presentation to MAIT cells. The MR1 promoter contains binding motifs for interferon regulatory factor 1 (IRF1), an important MHC-I transcription factor. IRF1 knockout reduced IFNγ-stimulated MR1 transcription, surface expression, and antigen presentation. Conversely, knockout of Nod-like Receptor family CARD domain-containing 5 (NLRC5), a critical component of MHC-I transcription, did not significantly impact MR1 expression. These findings were corroborated with IFNγ-treated primary AEC. MAIT cells in co-culture with Streptococcus pneumoniae-infected AEC produced sufficient IFNγ to stimulate MR1 expression.
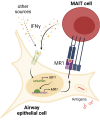
Conclusion: Our data support a model where IFNγ from activated MAIT cells or another source stimulates IRF1-dependent MR1 expression and antigen presentation, leading to greater MAIT cell activation. A robust MR1-dependent MAIT cell response may be beneficial for early infection responses, allowing minimal antigen stimulus to generate greater proinflammatory activity.
Keywords: IRF1; MAIT (mucosal-associated invariant T) cell; MR1; airway epithelial cell; interferon-gamma.
Copyright © 2025 Huber, Larson, Lust, Heisler and Harriff.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

