Mechanism by which porcine transmissible gastroenteritis virus disrupts host innate immunity
- PMID: 41080534
- PMCID: PMC12507764
- DOI: 10.3389/fimmu.2025.1675572
Mechanism by which porcine transmissible gastroenteritis virus disrupts host innate immunity
Abstract
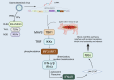
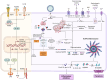
Innate immune evasion is a critical aspect of viral infections, as it disrupts the host's defense mechanisms.The innate immune system, as the primary defense against pathogens, detects pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRRs). This recognition triggers the production of interferons (IFNs) and pro-inflammatory factors, initiating the antiviral immune response. During evolution, viruses have found many ways to evade innate immune response in order to increase the replication efficiency, transmission ability and to establish persistent infection through co-evolution with hosts. Pigs act as natural hosts for a variety of significant viruses, including both DNA and RNA viruses. These viruses not only jeopardize animal health but also present a potential risk of interspecies transmission. Among these, porcine transmissible gastroenteritis virus (TGEV) stands out as a highly prevalent and severely detrimental enterovirus in the global swine industry. This review aims to comprehensively analyze the interaction between TGEV and host cells, emphasizing the molecular underpinnings of its immune evasion strategies. In addition, we will describe the programmed cell death types induced by TGEV, including autophagy, apoptosis and pyroptosis. Compared with existing reviews, this article not only provides a systematic integration of the multilayered immune evasion mechanisms of TGEV but also, for the first time, offers a comprehensive overview of its interactions with various forms of programmed cell death. This perspective highlights the complex regulatory networks underlying TGEV's adaptive evolution in the host, thereby enhancing our understanding of the pathogenic mechanisms of porcine coronaviruses and offering novel theoretical foundations for the development of vaccines and antiviral therapeutics.
Keywords: IFN; PAMPS; PRRs; TGEV; immune response; innate immune escape.
Copyright © 2025 Wang, Xie, Li, Liu, Zhang, Mi, Wang, Wang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

