HuR ablation destabilizes Foxp3 mRNA and impairs regulatory T cell function, contributing to an autoimmune phenotype
- PMID: 41080546
- PMCID: PMC12511036
- DOI: 10.3389/fimmu.2025.1618677
HuR ablation destabilizes Foxp3 mRNA and impairs regulatory T cell function, contributing to an autoimmune phenotype
Abstract
Introduction: The RNA-binding protein HuR (Elavl1), a key post-transcriptional regulator, plays a critical role in T cell activation and function by stabilizing target mRNAs. To investigate the role of HuR in regulatory T cell (Treg) function, we generated the Foxp3YFP/Cre HuRfl/fl mouse model.
Methods: In this model, homozygous females and hemizygous males for Foxp3 developed a scurfy-like phenotype displaying autoimmune features, including failure to thrive, splenomegaly, hair loss, tail stippling, and widespread multi-organ immune cell infiltration. Molecular analysis included direct interaction studies between HuR and Foxp3 mRNA to assess mRNA stability, RNA sequencing of YFP⁺ Tregs, Protein-Protein Interaction (PPI) analysis, qPCR, and Treg functional assays.
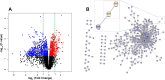
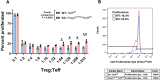
Results: To our knowledge, this is the first study demonstrating that HuR directly binds and stabilizes Foxp3 mRNA in Tregs, using a novel Treg-specific HuR-deficient mouse model, with implications for autoimmune regulation. Foxp3 mRNA stability and expression were significantly reduced in Tregs from these HuR KO mice, despite higher frequencies of YFP⁺ Tregs. RNA sequencing revealed significant dysregulation of several pathways, including the T helper differentiation pathway, in which Foxp3 played a central role. PPI analysis showed a direct link between Foxp3 and Rorc (encoding RORγt), connecting Foxp3 to the T cell differentiation pathway via IL-23R. Our qPCR analysis supported these findings. Functional assays demonstrated a reduction in the suppressive capacity of HuR-deficient Tregs.
Conclusion: These findings together suggest that ablation of HuR in Tregs disrupts Foxp3 expression and Treg function, likely through dysregulation of T cell differentiation pathways involving RORγt. This potentially contributes to a disrupted Treg-Th17 axis and autoimmune dysfunction. These data suggest that HuR-mediated post-transcriptional regulation contributes to maintaining Foxp3 expression and immune homeostasis, although compensatory mechanisms such as increased IL-10 expression may also be involved.
Keywords: Foxp3 stability; HuR (ELAVL1); IPEX syndrome; RNA-binding proteins (RBPs); RORγt (Rorc); post-transcriptional regulation; regulatory T cells (Tregs); systemic autoimmunity.
Copyright © 2025 Fattahi, Ellis, Vallance, Bahleda, Holden, Socha, Meier, Gomez-Rivera and Atasoy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

