Multi-omics reveal critical roles of phosphatidylcholine and sphingomyelin in antipsychotic efficacy for schizophrenia
- PMID: 41083443
- PMCID: PMC12518867
- DOI: 10.1038/s41392-025-02431-4
Multi-omics reveal critical roles of phosphatidylcholine and sphingomyelin in antipsychotic efficacy for schizophrenia
Abstract
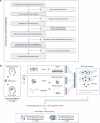
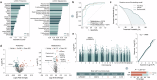
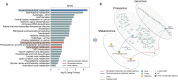
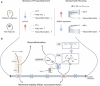
Nearly 30% of patients with schizophrenia respond inadequately to current antipsychotics, with unclear markers and mechanisms of antipsychotic efficacy. A total of 208 patients with schizophrenia treated for 6 weeks with oral paliperidone were analyzed through genotyping, mass spectrometry proteomic, and metabolomic profiling to explore underlying markers and mechanisms of antipsychotic efficacy. Machine learning analysis identified 20 proteins and 20 metabolites at baseline predictive of treatment response. Proteomic and metabolomic models achieved a cross-site mean AUC of 0.923 and 0.816, respectively. A multi-omics ensemble model achieved 0.941. GWAS and differential analyses identified 32 loci (P < 5 × 10-5), 83 proteins, and 31 metabolites associated with efficacy (P < 0.05). Trans-omics analysis of these efficacy-related molecules across three omic layers highlighted glycerophospholipid metabolism (P = 3.25 × 10-5) and sphingolipid metabolism (P = 0.039). Key molecules within these pathways exhibited a consistent direction of effect in regulating phosphatidylcholine (PC) and sphingomyelin (SM) metabolism, and higher PC and SM levels were found to correlate with better efficacy. These associations were further genetically validated using polygenic risk scores in two independent cohorts (2281 and 449 patients, respectively). In conclusion, multi-omics modeling is able to accurately identify antipsychotic efficacy, and higher PC and SM levels correlate with better antipsychotic efficacy, suggesting that variations in phospholipid metabolism may underlie the response to antipsychotics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- McCutcheon, R. A., Marques, T. R. & Howes, O. D. Schizophrenia—an overview. JAMA Psychiatry77, 201–210 (2020). - PubMed
-
- Demjaha, A. et al. Antipsychotic treatment resistance in first-episode psychosis: prevalence, subtypes and predictors. Psychol. Med.47, 1981–1989 (2017). - PubMed
-
- Pandurangi, A. K. & Buckley, P. F. Inflammation, antipsychotic drugs, and evidence for effectiveness of anti-inflammatory agents in schizophrenia. in Neuroinflammation and Schizophrenia Vol. 44 (eds Khandaker, G. M., Meyer, U. & Jones, P. B.) 227–244 (Springer International Publishing, 2019). - PubMed
MeSH terms
Substances
Grants and funding
- 82330042/National Natural Science Foundation of China (National Science Foundation of China)
- 82441005/National Natural Science Foundation of China (National Science Foundation of China)
- 82501802/National Natural Science Foundation of China (National Science Foundation of China)
- 82301687/National Natural Science Foundation of China (National Science Foundation of China)
- 82571710/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical

