Capturing disease severity in LIS1-lissencephaly reveals proteostasis dysregulation in patient-derived forebrain organoids
- PMID: 41083500
- PMCID: PMC12518603
- DOI: 10.1038/s41467-025-64980-0
Capturing disease severity in LIS1-lissencephaly reveals proteostasis dysregulation in patient-derived forebrain organoids
Abstract
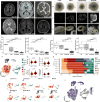
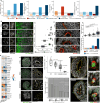
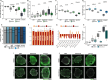
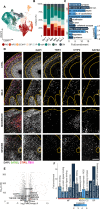
LIS1-lissencephaly is a neurodevelopmental disorder marked by reduced cortical folding and severe neurological impairment. Although all cases result from heterozygous mutations in the LIS1 gene, patients present a broad spectrum of severity. Here, we use patient-derived forebrain organoids representing mild, moderate, and severe LIS1-lissencephaly to uncover mechanisms underlying this variability. We show that LIS1 protein levels vary across patient lines and partly correlate with clinical severity, indicating mutation-specific effects on protein function. Integrated morphological, transcriptomic, and proteomic analyses reveal progressive changes in neural progenitor homeostasis and neurogenesis that scale with severity. Mechanistically, microtubule destabilization disrupts cell-cell junctions and impairs WNT signaling, and defects in protein homeostasis, causing stress from misfolded proteins, emerge as key severity-linked pathways. Pharmacological inhibition of mTORC1 partially rescues these defects. Our findings demonstrate that patient-derived organoids can model disease severity, enabling mechanistic dissection and guiding targeted strategies in neurodevelopmental disorders.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: O. F. has affiliated with Springer Nature following the completion of her work on this project. The other authors have no relevant financial or non-financial interests to disclose.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

