A molecular circuit regulates fate plasticity in emerging and adult AT2 cells
- PMID: 41087371
- PMCID: PMC12521654
- DOI: 10.1038/s41467-025-64224-1
A molecular circuit regulates fate plasticity in emerging and adult AT2 cells
Abstract
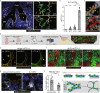
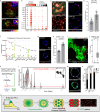
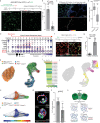
Alveolar Type 1 and Type 2 cells are vital for lung gas exchange and become compromised in several diseases. While key differentiation signals are known, their emergence and fate plasticity are unclear. Here we show in the embryonic lung that single AT2s emerge at intermediate zones, extrude, and connect with nearby epithelium via interlumenal junctioning. We observe AT2s retain fate plasticity until the bZIP transcription factor C/EBPα suppresses Notch signaling at a novel Dlk1 enhancer. Both Dlk1 and Cebpa are regulated by the polycomb repressive complex (PRC2), which together form a "pulse generator" circuit that times Dlk1 expression and thus Notch activation, resulting in a "salt and pepper" pattern of AT1 and AT2 fate. In injured adult lungs, C/EBPα downregulation is required to re-access AT2 fate plasticity and is mediated by the dominant negative C/EBP family member CHOP. Finally, Cebpa loss also activates a "defender" AT2 state, distinct from its reparative state, and we propose AT2s toggle between either state following infection to protect and repair alveoli.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Update of
-
A molecular circuit regulates fate plasticity in emerging and adult AT2 cells.bioRxiv [Preprint]. 2025 May 7:2025.04.28.650846. doi: 10.1101/2025.04.28.650846. bioRxiv. 2025. Update in: Nat Commun. 2025 Oct 14;16(1):8924. doi: 10.1038/s41467-025-64224-1. PMID: 40654759 Free PMC article. Updated. Preprint.
References
-
- Barnes, P. J. et al. Chronic obstructive pulmonary disease. Nat. Rev. Dis. Prim.1, 15076 (2015). - PubMed
-
- Martinez, F. J. et al. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Prim.3, 17074 (2017). - PubMed
-
- Siegel, R., Naishadham, D. & Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin.63, 11–30 (2013). - PubMed
MeSH terms
Substances
Grants and funding
- R00 HL127267/HL/NHLBI NIH HHS/United States
- R01 HL171056/HL/NHLBI NIH HHS/United States
- R01HL171056/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R00HL127267/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

