A novel six-biomarker panel identified from male breast cancer-associated fibroblasts demonstrates prognostic power for prostate tumors
- PMID: 41088372
- PMCID: PMC12522489
- DOI: 10.1186/s12967-025-07196-6
A novel six-biomarker panel identified from male breast cancer-associated fibroblasts demonstrates prognostic power for prostate tumors
Abstract
Background: Male breast cancer (BC) is rare, accounting for only approximately 1% of all cases of BC, and poorly characterized. In contrast, prostate cancer is the most prevalent cancer in men and serves as a model for understanding male-specific tumor biology. The advent of high-throughput technologies has enabled the development of gene expression signatures for both breast and prostate tumors that could inform prognosis and guide treatment. In this respect, the role of the tumor microenvironment, particularly cancer-associated fibroblasts (CAFs), remains largely underexplored. Here, we sought to identify a CAF-related gene signature in male patients with BC and prostate cancer to reveal specific protumorigenic mechanisms and identify novel therapeutic targets for both malignancies.
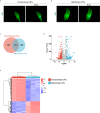
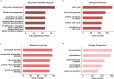
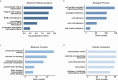
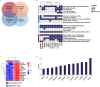
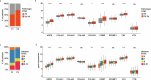
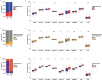
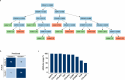
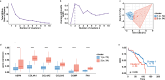
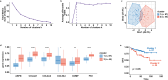
Methods: RNA sequencing was performed to analyze and compare the transcriptomes of CAFs isolated from female and male BC patients. Differentially expressed genes (DEGs) between female and male breast CAFs were identified and subjected to enrichment analyses. Using a set of candidate upregulated genes in male breast CAFs, K-means clustering of prostate cancer patients was performed using multiple datasets to define a prognostic gene signature. Kaplan‒Meier curves and log-rank tests were conducted to assess differences in patient outcomes and other clinical variables between groups of patients with high or low prognostic gene expression. The clustering results were then validated using decision tree analysis, and boosted calculations were employed to increase the classifier performance.
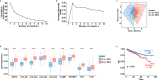
Results: Transcriptomic profiling revealed 775 DEGs between female and male breast CAFs. Owing to the limited transcriptomic data from male BC patients, we leveraged large prostate cancer cohorts to investigate the relevance of the genes expressed by male breast CAFs. A six-gene signature (ASPN, COL4A1, COL4A2, COL5A3, COMP and FN1) that could predict patient outcomes in multiple independent cohorts of prostate cancer patients was identified.
Conclusions: We identified a novel gene signature with strong prognostic value in prostate cancer and potential relevance to male BC. This gene signature represents a complementary tool to standard clinical parameters for improving patient stratification and management.
Keywords: Cancer-associated fibroblasts (CAFs); Gene signature; K-means; Male breast cancer; Prostate cancer; RNA-sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All procedures conformed to the Helsinki Declaration for the research on humans. Signed informed consent was obtained from all patients and the experimental research has been performed with the ethical approval provided by the “Comitato Etico Regione Calabria, Cosenza, Italy” (approval code: 500/2022). Consent for publication: Not applicable. Competing interests: RL is Associate Editor. MM is Section Editor.
Figures

References
-
- Kim J, Harper A, McCormack V, Sung H, Houssami N, Morgan E, et al. Global patterns and trends in breast cancer incidence and mortality across 185 countries. Nat Med. 2025;31:1154–62. - PubMed
-
- Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primers. 2019;5:66. - PubMed
-
- Ray SK, Mukherjee S. Clinical aspect of male breast cancer: a burgeoning and unaddressed issue. Mol Biol Rep. 2025;52:452. - PubMed
-
- Bhardwaj PV, Gupta S, Elyash A, Teplinsky E. Male breast cancer: A review on diagnosis, treatment, and survivorship. Curr Oncol Rep. 2024;26:34–45. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

