FASN inhibits ferroptosis in breast cancer via USP5 palmitoylation-dependent regulation of GPX4 deubiquitination
- PMID: 41088402
- PMCID: PMC12523187
- DOI: 10.1186/s13046-025-03548-8
FASN inhibits ferroptosis in breast cancer via USP5 palmitoylation-dependent regulation of GPX4 deubiquitination
Abstract
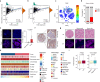
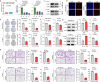
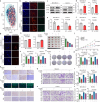
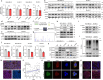
Increasing studies have reported that dysregulated lipid metabolism is an independent risk factor for breast cancer (BC); it would be, therefore, enlightening to investigate the relationship between metabolic reprogramming and the tumor microenvironment in the future. Ferroptosis, a novel form of programmed cell death, is characterized by glutathione (GSH) depletion and inactivation of glutathione peroxidase 4 (GPX4), the central regulator of the antioxidant system. While the close association between fatty acid metabolism and ferroptosis has been studied in various diseases, the interplay between the key fatty acid metabolic enzyme fatty acid synthase (FASN) and ferroptosis in BC remains unexplored. At the beginning of the current study, we demonstrated that FASN expression positively correlates with an immune-cold tumor microenvironment in BC. Subsequent findings revealed that FASN knockdown promotes GPX4 degradation-induced ferroptosis, thereby enhancing the efficacy of anti-programmed cell death protein 1 (PD-1) immunotherapy. Co-immunoprecipitation coupled with mass spectrometry (IP/MS) and co-IP experiments demonstrated that ubiquitin specific protease 5 (USP5) stabilizes GPX4 by binding to and deubiquitinating it. Furthermore, knockdown of FASN inhibited the palmitoylation of USP5, reducing its interaction with GPX4 and consequently increasing GPX4 ubiquitination and degradation. Our results demonstrate that FASN suppresses ferroptosis in BC by stabilizing GPX4 via USP5-mediated mechanisms, highlighting FASN inhibition as a potential therapeutic approach to enhance immunotherapy response.
Keywords: Breast cancer; FASN; Ferroptosis; Palmitoylation; USP5.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study involves human participants and ethical approval for the study of TMAs was granted by the Outdo Biotech Clinical Research Ethics Committee. Ethical approval for the single-cell sequencing data was granted by Clinical Research Ethics Committees at Wuxi Maternal and Child Health Care Hospital (Ethics No. 2024–01-1024–31). Participants gave informed consent to participate in the study before taking part. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Jiangnan University (JN. No 20250228b0720731[069]). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
-
- Bray F, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Loibl S, et al. Early breast cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann Oncol. 2024;35:159–82. - PubMed
-
- Gradishar WJ, et al. Breast cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20:691–722. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

