Circulating endothelial signatures correlate with worse outcomes in COVID-19, respiratory failure and ARDS
- PMID: 41088445
- PMCID: PMC12522733
- DOI: 10.1186/s13054-025-05596-0
Circulating endothelial signatures correlate with worse outcomes in COVID-19, respiratory failure and ARDS
Abstract
Background: Elevated circulating endothelial cells (CECs), released from monolayers after insult, have been implicated in worse outcomes in ARDS and COVID-19, however there is no consensus proteomic phenotype that define CECs. We queried whether a transcriptomic approach would alternatively support the presence of endothelial cells in circulation and correlate with worsening respiratory failure.
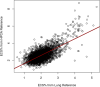
Methods: To test whether elevated endothelial cell signatures (ECS) in circulation plays a role in worse respiratory outcomes, we used unsupervised bulk-transcriptome deconvolution to quantify ECS% in two cohorts. Our pilot analysis included pediatric patients requiring invasive mechanical ventilation (CAF-PINT, NCT01892969). Our validation cohort included adult hospitalized patients with COVID-19 (IMPACC, NCT04378777), testing the association of ECS% to outcomes in patients at risk of acute respiratory failure/ARDS. Primary outcome was 28-day mortality.
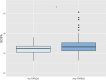
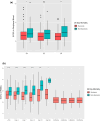
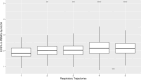
Results: In CAF-PINT, day 0 ECS% was higher in non-survivors compared to survivors of respiratory failure (2.8%, IQR 2.4-3.4% versus 2.6%, IQR 2.2-3.0% n = 244, p < 0.05, Wilcoxon rank-sum). In IMPACC, baseline ECS% (< 72 h of hospitalization) was higher in COVID-19 non-survivors versus survivors (2.9%, IQR 2.6-3.4%, versus 2.7%, IQR 2.3-3.1%, n = 932, p < 0.001, Wilcoxon rank-sum). Each 1% increase in baseline ECS% was significantly associated with mortality (adjusted OR 1.36, CI 1.03-1.79) by multivariable logistic regression. Increased baseline ECS% was associated with worse respiratory trajectories (2.5%, IQR 2.2-2.8% for trajectory with no oxygen requirements, 2.9%, IQR 2.6-3.4% for the trajectory with fatal outcome by day 28, n = 932, p < 0.001, one-way ANOVA).
Conclusion: Quantifying ECS by deconvolution supports a transcriptomics-driven approach towards the non-invasive evaluation of endothelial damage in respiratory outcomes. This is a first step towards elucidating mechanistic components linking endothelial damage to ARDS utilizing non-invasive, circulating transcriptomic data by leveraging a novel deconvolution approach.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: CAFPINT/HAFPINT- The study was coordinated by a Clinical Coordinating Center (CCC) with staff from both CCC Principal Investigators’ sites (PIs MA, VN) and a Data Coordinating Center (DCC; PI DW, U01 HL108028). Patients were screened and recruited at 35 sites in the United States, Canada, and Australia. Central ethics review was arranged at the Boston Children’s Hospital Institutional Review Board (IRB) to be available for sites willing to cede ethics review authority with appropriate signed institutional agreements. Ten sites established this reliance relationship, while the remaining sites conducted local IRB review and oversight. Study protocol modifications were developed collaboratively between the study’s ten-member leadership committee and a steering committee comprised of co-investigators from all participating sites. Families of potential study patients were approached by the local study team upon confirmation of hyperglycemia [two consecutive BGs of ≥ 150 mg/dL (8.3 mmol/L)] after obtaining permission from the attending physician to approach the family. Attending physicians were asked to confirm that eligible female patients were not pregnant prior to their families being approached to minimize any risk to the fetus from study-induced hypoglycemia. An informed consent and/or assent discussion with the family and/or patient took place in the ICU or in a nearby private room. Patients whose families declined participation in the study were managed according to usual practice by the local clinical team, which may or may not have included therapy for glycemic control. Consented subjects with confirmed hyperglycemia were randomized to one of the two treatment arms. IMPACC: NIAID staff conferred with the Department of Health and Human Services Office for Human Research Protections (OHRP) regarding the potential applicability of the public health surveillance exception [45CFR46.102(l) (2)] to the IMPACC study protocol. OHRP concurred that the study satisfied criteria for the public health surveillance exception, and the IMPACC study team sent the study protocol, and participant information sheet for review and assessment to institutional review boards (IRBs) at participating institutions. Twelve institutions elected to conduct the study as public health surveillance, while 3 sites with prior IRB-approved biobanking protocols elected to integrate and conduct IMPACC under their institutional protocols (University of Texas at Austin, IRB 2020-04-0117; University of California San Francisco, IRB 20-30497; Case Western Reserve University, IRB STUDY20200573) with informed consent requirements. Participants enrolled under the public health surveillance exclusion were provided information sheets describing the study, samples to be collected, and plans for data deidentification and use. Those who requested not to participate after reviewing the information sheet were not enrolled. In addition, participants did not receive compensation for study participation while inpatient, and subsequently were offered compensation during outpatient follow-ups. Consent for publication: All authors have read the manuscript and have consented to its publication. Competing interests: The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines which list Florian Krammer as co-inventor. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. Florian Krammer has consulted for Merck and Pfizer (before 2020), and is currently consulting for Pfizer, Seqirus, 3rd Rock Ventures, Merck and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2. Viviana Simon is a co-inventor on a patent filed relating to SARS-CoV-2 serological assays (the “Serology Assays”). Ofer Levy is a named inventor on patents held by Boston Children’s Hospital relating to vaccine adjuvants and human in vitro platforms that model vaccine action. His laboratory has received research support from GlaxoSmithKline (GSK) and he has served as a consultant for GSK and Hillevax. He is also a co-founder of and advisor to Ovax, Inc, a company developing vaccines against opioid overdose. Charles Cairns serves as a consultant to bioMerieux and is funded for a grant from Bill & Melinda Gates Foundation. James A Overton is a consultant at Knocean Inc. Jessica Lasky-Su serves as a scientific advisor of Precion Inc. Scott R. Hutton, Greg Michelloti and Kari Wong are employees of Metabolon Inc. Vicki Seyfer- Margolis is a current employee of MyOwnMed. Emory University receives funds for Nadine Rouphael to conduct research from Sanofi, Lilly, Merck, Quidel, Immorna, Vaccine Company and Pfizer. Nadine Rouphael served on selected advisory boards for Sanofi, Seqirus, Pfizer and Moderna and is a paid clinical trials safety consultant for ICON, CyanVac, Imunon and EMMES. Adeeb Rahman is a current employee of Immunai Inc. Steven Kleinstein is a consultant related to ImmPort data repository for Peraton. Nathan Grabaugh is a consultant for Tempus Labs and the National Basketball Association. Akiko Iwasaki is a consultant for 4BIO, Blue Willow Biologics, Revelar Biotherapeutics, RIGImmune, Xanadu Bio, Paratus Sciences. Monika Kraft receives research funds paid to her institution from NIH, ALA; Sanofi, Astra-Zeneca for work in asthma, serves as a consultant for Astra-Zeneca, Sanofi, Chiesi, GSK for severe asthma; is a co-founder and CMO for RaeSedo, Inc, a company created to develop peptidomimetics for treatment of inflammatory lung disease. Esther Melamed received research funding from Babson Diagnostics, honorarium from Multiple Sclerosis Association of America and has served on advisory boards of Genentech, Horizon, Teva and Viela Bio. Carolyn Calfee receives research funding from NIH, FDA, DOD, Roche-Genentech and Quantum Leap Healthcare Collaborative as well as consulting services for Janssen, Vasomune, Gen1e Life Sciences, NGMBio, and Cellenkos. Wade Schulz was an investigator for a research agreement, through Yale University, from the Shenzhen Center for Health Information for work to advance intelligent disease prevention and health promotion; collaborates with the National Center for Cardiovascular Diseases in Beijing; is a technical consultant to Hugo Health, a personal health information platform; cofounder of Refactor Health, an AI-augmented data management platform for health care; and has received grants from Merck and Regeneron Pharmaceutical for research related to COVID-19. Grace A McComsey received research grants from Rehdhill, Cognivue, Pfizer, and Genentech, and served as a research consultant for Gilead, Merck, Viiv/GSK, and Jenssen. Linda N. Geng received research funding paid to her institution from Pfizer, Inc.
Figures

References
-
- Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med. 2000;342:1334–49. - PubMed
-
- Wick KD, et al. Opportunities for improved clinical trial designs in acute respiratory distress syndrome. Lancet Respir Med. 2022;10:916–24. - PubMed
-
- Woywodt A, Bahlmann FH, de Groot K, Haller H, Haubitz M. Circulating endothelial cells: life, death, detachment and repair of the endothelial cell layer. Nephrol Dial Transplant. 2002;17:1728–30. - PubMed
MeSH terms
Grants and funding
- U19 AI118608/AI/NIAID NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- R01 HL114484/HL/NHLBI NIH HHS/United States
- U19AI128910/NH/NIH HHS/United States
- U19 AI062629/AI/NIAID NIH HHS/United States
- U01HL107681/HL/NHLBI NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- S10 OD026799/OD/NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- R01 HL170601/HL/NHLBI NIH HHS/United States
- U24AI52179/NH/NIH HHS/United States
- R01-HL166449, R01-HL169860/HL/NHLBI NIH HHS/United States
- U19AI118608/NH/NIH HHS/United States
- U19AI062629/NH/NIH HHS/United States
- U19AI057229/NH/NIH HHS/United States
- U19 AI090023/AI/NIAID NIH HHS/United States
- U19 AI125357/AI/NIAID NIH HHS/United States
- R01 AI145835/AI/NIAID NIH HHS/United States
- U19AI090023/NH/NIH HHS/United States
- U19 AI128913/AI/NIAID NIH HHS/United States
- R01 AI132774/AI/NIAID NIH HHS/United States
- R01AI145835/NH/NIH HHS/United States
- U19AI077439/NH/NIH HHS/United States
- R01 HL166449/HL/NHLBI NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- R01AI135803/NH/NIH HHS/United States
- K23HL165149/HL/NHLBI NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U54AI142766/NH/NIH HHS/United States
- U19AI089992/NH/NIH HHS/United States
- U01 HL107681/HL/NHLBI NIH HHS/United States
- R01HL114484/NH/NIH HHS/United States
- R01AI132774/NH/NIH HHS/United States
- R01 HL169860/HL/NHLBI NIH HHS/United States
- R01 AI135803/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
- R01AI104870/NH/NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- U19AI118610/NH/NIH HHS/United States
- U19AI128913/NH/NIH HHS/United States
- R01 AI104870/AI/NIAID NIH HHS/United States
- R01HL170601/HL/NHLBI NIH HHS/United States
- K23 HL165149/HL/NHLBI NIH HHS/United States
- U19AI125357/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

