Cell line-specific estrogen responses uncover functional sex differences in murine macrophages
- PMID: 41088453
- PMCID: PMC12522439
- DOI: 10.1186/s13293-025-00760-1
Cell line-specific estrogen responses uncover functional sex differences in murine macrophages
Abstract
Background: RAW 264.7 (male-derived) and J774A.1 (female-derived) cell lines are widely used in immunology research and are considered preferred models for studying signaling pathways, yet their responses to gonadal hormones remain poorly understood. Gonadal hormones, particularly estrogen, shape immune cell function and contribute to sex differences in disease outcomes, with macrophages playing a central role through their expression of intracellular estrogen receptors (ERs). Herein, we investigated ER expression and functional responses to 17β-estradiol (E2) in male-derived RAW 264.7 and female-derived J774A.1 macrophages, in 2D culture. Additionally, we looked at sex-matched and mismatched media conditions in a 3D hydrogel system. Our results reveal distinct phenotypic and functional differences between the cell lines, emphasizing the need for sex-aware approaches in immunological research and model design.
Methods: RAW 264.7 and J774A.1 macrophages were cultured in basal media for 24 hours, then treated with varying concentrations of 17β-estradiol (5, 25, 100 nM), as well as hormone-free and control media. Post-treatment analyses included viability, estrogen receptor expression, phenotype skewing, matrix metalloprotease 9 (MMP9) activity, and phagocytosis. These macrophages were also used to condition sex-specific media environments and were encapsulated in a hydrogel network containing adhesive and cleavable sites. Encapsulated cells were then exposed to sex-matched or sex-mismatched conditioned media, and proliferation and MMP9 activity were assessed.
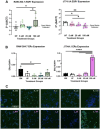
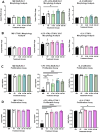
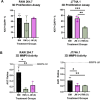
Results: Our results revealed distinct differences in estrogen receptor gene and protein expression, as well as in core macrophage functions such as proliferation, inflammation, matrix remodeling, and phenotype skewing. Additionally, the sex-derivation of the surrounding molecular environment affected macrophage behavior in a 3D hydrogel system. Female-derived macrophages were more sensitive in terms of proliferation to sex-mismatched environments, while male-derived macrophages exhibited altered enzyme activity when exposed to female-conditioned media.
Conclusions: These findings underscore the importance of accounting for both the origin of immune cells as well as the hormonal and environmental context in which they are studied. Without these considerations, experimental models risk missing critical biological differences that shape immune responses and disease outcomes.
Keywords: Estrogen; Gonadal hormones; Macrophage function.
Plain language summary
Males and females often experience different symptoms, risks, and outcomes when it comes to certain diseases and health conditions. One reason for this may be that male and female immune cells behave differently, especially in response to hormones like estrogen. In this study, we looked at two commonly used types of mouse immune cells—one originally from a male and one from a female—to see how they respond to estrogen.We found that male and female cells do not respond to estrogen in the same way. They showed different levels of activity, growth, and behavior depending on both the hormone exposure and the sex origin of the environment they were in. We also placed the cells in a gel that mimics tissue and exposed them to sex-specific environments and saw clear differences in how male and female cells responded. For example, female-derived cells were more sensitive in their ability to grow when placed in a “male” environment, while male-derived cells changed their behavior when exposed to signals from a “female” environment.Our findings show that both the origin of immune cells and the environment they are placed in can strongly influence how they behave. This means that to better understand immune responses and develop more effective treatments, scientists need to consider sex as a key factor in their research models. Ignoring these differences could lead to incomplete or misleading results.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Update of
-
Cell Line-Specific Estrogen Responses Uncover Functional Sex Differences in Murine Macrophages.Res Sq [Preprint]. 2025 Jun 30:rs.3.rs-6925474. doi: 10.21203/rs.3.rs-6925474/v1. Res Sq. 2025. Update in: Biol Sex Differ. 2025 Oct 14;16(1):75. doi: 10.1186/s13293-025-00760-1. PMID: 40630528 Free PMC article. Updated. Preprint.
References
-
- Campbell L, Emmerson E, Williams H, Saville CR, Krust A, Chambon P, et al. Estrogen receptor-alpha promotes alternative macrophage activation during cutaneous repair. J Invest Dermatol. 2014;134(9):2447–57. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

