A flexible systems analysis pipeline for elucidating spatial relationships in the tumor microenvironment linked with cellular phenotypes and patient-level features
- PMID: 41089681
- PMCID: PMC12515857
- DOI: 10.3389/fimmu.2025.1642527
A flexible systems analysis pipeline for elucidating spatial relationships in the tumor microenvironment linked with cellular phenotypes and patient-level features
Abstract
Introduction: Quantitative investigation of how the spatial organization of cells within the tumor microenvironment associates with disease progression, patient outcomes, and that cell's phenotypic state remains a key challenge in cancer biology. High-dimensional multiplexed imaging offers an opportunity to explore these relationships at single-cell resolution.
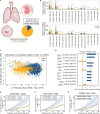
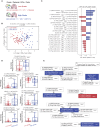
Methods: We developed a computational pipeline to quantify and analyze the neighborhood profiles of individual cells in multiplexed immunofluorescence images. The pipeline characterizes spatial co-localization patterns within the tumor microenvironment and applies interpretable supervised machine learning models, specifically orthogonal partial least squares analysis (OPLS), to identify spatial relationships predictive of cell states and clinical phenotypes.
Results: We applied this framework to a previously published non-small cell lung cancer (NSCLC) cohort across four applications. At the cellular level, we identified neighborhood features associated with lymphocyte activation states. At the tumor-immune interface, we demonstrated that the immune cell composition surrounding major histocompatibility complex class I-expressing (MHC I+) tumor cells could distinguish adenocarcinoma from squamous cell carcinoma. At the patient level, spatial features predicted tumor grade.
Discussion: By integrating cell-segmented imaging data with interpretable modeling, our pipeline reveals key spatial determinants of tumor biology. These findings generate testable mechanistic hypotheses about intercellular interactions and support the development of spatially informed prognostic and therapeutic strategies.
Keywords: NK cell; T cell; immune interactions; spatial biology; spatial proteomics; supervised machine learning; systems immunology; tumor-immune cell interactions.
Copyright © 2025 Hanson, Goundry, Wessel, Brown, Bullock and Dolatshahi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures




References
-
- Wu M-Y, Lu J-H. Autophagy and macrophage functions: inflammatory response and phagocytosis. Cells. (2020) 9:70. Available online at: https://www.mdpi.com/2073-4409/9/1/70. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

