The Taiwan Precision Medicine Initiative provides a cohort for large-scale studies
- PMID: 41092961
- PMCID: PMC12675286
- DOI: 10.1038/s41586-025-09680-x
The Taiwan Precision Medicine Initiative provides a cohort for large-scale studies
Abstract
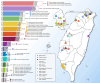
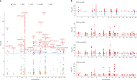
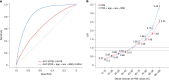
Han Chinese people comprise nearly 20% of the global population but remain under-represented in genetic studies1,2, so there is an urgent need for large-scale cohorts to advance precision medicine. Here we present the Taiwan Precision Medicine Initiative (TPMI), established by Academia Sinica in collaboration with 16 major medical centres around Taiwan, which has recruited 565,390 participants who consent to provide DNA samples for genetic profiling and grant access to their electronic medical records (EMRs) for research. EMR access is both retrospective and prospective, allowing longitudinal studies. Genetic profiling is done with population-optimized arrays of single-nucleotide polymorphisms for people of Han Chinese ancestry, which enable genome-wide association3,4, phenome-wide association5,6 and polygenic risk score7,8 studies to be performed to evaluate common disease risk and pharmacogenetic response. Participants also agreed to be re-contacted for future research and receive personalized genetic risk profiles with health management recommendations. The TPMI has established the TPMI Data Access Platform, a central database and analysis platform that both safeguards the security of the data and facilitates academic research. As a large cohort of individuals with non-European ancestry that merges genetic profiles with EMR data and enables longitudinal follow-up, TPMI provides a unique resource that could be used to validate genetic risk prediction models, perform clinical trials of risk-based health management and inform health policies. Ultimately, the TPMI cohort will contribute to global genetic research and serve as a model for population-based precision medicine.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Primers1, 59 (2021).
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

