Low circulating miR-190a-5p predicts progression of chronic kidney disease
- PMID: 41093838
- PMCID: PMC12528679
- DOI: 10.1038/s41467-025-64168-6
Low circulating miR-190a-5p predicts progression of chronic kidney disease
Abstract
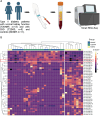
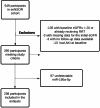
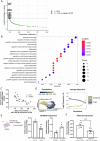
MicroRNAs may act as diagnostic and prognostic biomarkers of chronic kidney disease and are functionally important in disease pathogenesis. To identify novel microRNA biomarkers, we performed small RNA-sequencing on plasma from individuals with type 2 diabetes, with and without chronic kidney disease. MiR-190a-5p abundance was significantly lower in the circulation of type 2 diabetic patients with reduced function compared to those with normal kidney function. In an independent cohort of patients with chronic kidney disease of diverse aetiology, miR-190a-5p abundance predicted disease progression in individuals with no or moderate albuminuria ( < 300 mg/mmol). miR-190a-5p expression in kidney biopsy tissue correlated with the level of miR-190a-5p in the circulation and with estimated glomerular filtration rate, tubular mass and negatively with histological fibrosis. Administration of a miR-190a-5p mimic in a murine ischaemia-reperfusion injury model in male mice reduced tubular injury and fibrosis and increased expression of genes associated with tubular health. Our analyses suggest that miR-190a-5p is a biomarker of tubular cell health, low circulating levels may predict chronic kidney disease progression independent of existing risk factors and strategies to preserve miR-190a-5p may be an effective treatment for restoring tubular cell health following kidney injury.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: These authors declare no competing interests: J.Z., K.L.C., O.T., R.K.Y.W., M.R., C.C., J.P.T., J.H., A.P.M., G.J.M., D.A.S. and L.D. These authors declare the following competing interests: P.B.M. reports lecture or advisory board honoraria from AstraZeneca, Bayer, Boehringer Ingelheim, Pharmacosmos, Astellas, Vifor, GSK and research funding from AstraZeneca and Boehringer Ingelheim outside the submitted work. B.R.C. reports consultancy from Argenx and research funding from AstraZeneca outside the submitted work. D.P.B. is listed as a co-inventor on a patent application covering urinary protein biomarkers in chronic kidney disease outside the submitted work.
Figures



References
-
- Magliano, D. J., Boyko, E. J. IDF Diabetes Atlas. (2021).
-
- UK Renal Registry. UK Renal Registry 25th Annual Report – data to 31/12/2021 B, UK. https://ukkidney.org/audit-research/annual-report (2023).
-
- Johansen, K. L. et al. US Renal Data System 2023 Annual Data Report: Epidemiology of Kidney Disease in the United States. AJKD 83, A8–A13 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

