Piezo1 regulates the mechanotransduction of soft matrix viscoelasticity
- PMID: 41093846
- PMCID: PMC12528374
- DOI: 10.1038/s41467-025-64185-5
Piezo1 regulates the mechanotransduction of soft matrix viscoelasticity
Abstract
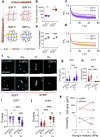
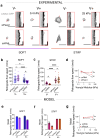
Mechanosensitive ion channels such as Piezo1 have fundamental roles in sensing the mechanical properties of the extracellular matrix. However, whether and how Piezo1 senses time-dependent matrix mechanical properties, that is, viscoelasticity, remains unknown. To address this question, we combine an immortalised mesenchymal stem cell line, in which Piezo1 expression can be silenced, with soft and stiff viscoelastic hydrogels that have independently tuneable elastic and viscous moduli. We demonstrate that Piezo1 is a regulator of the mechanotransduction of viscoelasticity in soft matrices, both experimentally and through simulations incorporating Piezo1 into a modified viscoelastic molecular clutch model. Using RNA sequencing, we also identify the transcriptomic responses of mesenchymal stem cells to matrix viscoelasticity and Piezo1 activity, identifying gene signatures that reflect their mechanobiology in soft and stiff viscoelastic hydrogels.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell126, 677–689 (2006). - PubMed
-
- Chan, C. E. & Odde, D. J. Traction dynamics of filopodia on compliant substrates. Science322, 1687–1691 (2008). - PubMed
-
- Elosegui-Artola, A., Trepat, X. & Roca-Cusachs, P. Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol.28, 356–367 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- 204820/Z/16/Z/Wellcome Trust (Wellcome)
- 101054728/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 101097753/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- EP/X033554/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
- 21-0156/AICR_/Worldwide Cancer Research/United Kingdom
LinkOut - more resources
Full Text Sources

