Nuclear receptor Nr1d1 links sleep deprivation to intestinal homeostasis via microbiota-derived taurine
- PMID: 41094491
- PMCID: PMC12522324
- DOI: 10.1186/s12967-025-07089-8
Nuclear receptor Nr1d1 links sleep deprivation to intestinal homeostasis via microbiota-derived taurine
Abstract
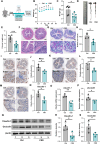
Background: Sleep deficiency significantly compromises human health, with the gut being particularly susceptible. However, the molecular mechanisms by which gut microbiota mediate sleep deprivation-induced intestinal dysfunction remain largely undefined. In this study, we employed a chronic sleep deprivation (CSD) mouse model to investigate the impact of sleep loss on intestinal integrity and microbial composition.
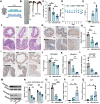
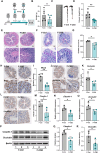
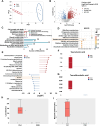
Methods: The CSD mouse model was established using the modified multiple platform (rotating rod) method. Colon histomorphology was assessed by hematoxylin and eosin (HE) staining. Expression levels of barrier proteins (Occludin, Claudin-1) and circadian regulators (Nr1d1, Bmal1) were evaluated via Western blot or immunohistochemistry (IHC). Gut microbiota composition and stability were analyzed by 16S rRNA gene sequencing, and the causal role of microbiota in CSD-induced barrier damage was assessed through fecal microbiota transplantation (FMT). RNA sequencing (RNA-seq) of intestinal epithelial tissues identified differentially expressed genes and enriched pathways. Untargeted metabolomics was employed to investigate key differential metabolites (Taurine). Additionally, taurine was supplemented in vivo to explore its efficacy and mechanism in alleviating intestinal barrier damage in CSD mice.
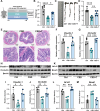
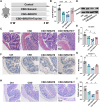
Results: CSD led to pronounced colon shortening and significant downregulation of the epithelial barrier proteins Occludin and Claudin-1, indicative of impaired intestinal barrier function. Moreover, CSD exacerbated symptoms of chemically induced colitis and induced gut microbiota dysbiosis. Mechanistically, FMT from CSD mice into antibiotic-treated recipients recapitulated intestinal inflammation, confirming the pathogenic role of the altered microbiota. Transcriptomic analysis revealed significant enrichment of genes involved in circadian rhythm pathways, notably a marked suppression of the circadian nuclear receptor Nr1d1, a key regulator of intestinal homeostasis. Complementary untargeted metabolomic profiling identified taurine as a microbiota-derived metabolite significantly reduced by CSD. In vivo taurine supplementation restored Nr1d1 expression, reinforced epithelial barrier integrity, and decreased pro-inflammatory cytokine production.
Conclusion: Together, these findings reveal a gut microbiota-taurine-Nr1d1 axis underlying sleep deprivation-induced intestinal barrier dysfunction, and suggest that therapeutic modulation of taurine levels or circadian pathways may offer novel strategies to prevent or treat sleep-related gastrointestinal disorders.
Keywords: Circadian rhythm; Gut microbiota; Nuclear receptor; Sleep deprivation; Taurine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing financial interests.
Figures


References
-
- Liew SC, Aung T. Sleep deprivation and its association with diseases—a review. Sleep Med. 2021;77:192–204. 10.1016/j.sleep.2020.07.048. - PubMed
-
- Lindsay S, Caroline W, Edwin WC, Aaron DF. Effect of sleep restriction on adolescent cognition by adiposity. JAMA Neurol. 2024. 10.1001/jamaneurol.2024.1332.
-
- Di S, et al. Prolonged sleep deprivation induces a cytokine-storm-like syndrome in mammals. Cell. 2023. 10.1016/j.cell.2023.10.025. - PubMed
MeSH terms
Substances
Grants and funding
- 2023YFC2705400/National Key R&D Rrogram of China
- 2021B1515420004/Guangdong Basic and Applied Basic Research Foundation
- 2025A03J4164/Huadu District Basic and Applied Basic Research Joint Funding Project
- GKLBCN-202501-01/Open Project Program of Guangxi Key Laboratory of Brain and Cognitive Neuroscience, Guilin Medical University
- 202201020363/Science and Technology Projects in Guangzhou
LinkOut - more resources
Full Text Sources

