Epigenetic profiles of tissue informative CpGs inform ALS disease status and progression
- PMID: 41094691
- PMCID: PMC12529837
- DOI: 10.1186/s13073-025-01542-5
Epigenetic profiles of tissue informative CpGs inform ALS disease status and progression
Abstract
Background: Cell-free DNA (cfDNA), derived from dying cells, has demonstrated utility across multiple clinical applications. However, its potential in neurodegenerative diseases remains underexplored, with most existing cfDNA technologies tailored to specific disease contexts like cancer or non-invasive prenatal screening.
Methods: To address this gap, we developed a novel approach to characterize epigenetic cfDNA profiles by identifying key regions of DNA methylation that reveal the tissues origins undergoing apoptosis or necrosis. We evaluated this method in the largest cfDNA study of amyotrophic lateral sclerosis (ALS) and other neurological diseases (OND) to date, encompassing two independent cohorts (n = 192) from Australia (UQ Ncases = 48, Ncontrols = 32, NOND = 15) and the USA, (UCSF Ncases = 50, Ncontrols = 45)).
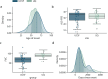
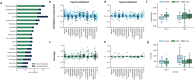
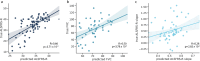
Results: Our approach accurately distinguished ALS patients from controls (UQ AUC = 0.82, UCSF AUC = 0.99) and from individuals with other neurological diseases (AUC = 0.91). It also identified an asymptomatic carrier of a pathogenic C9orf72 variant, and strongly correlated with ALS disease progression measures (Pearson's R = 0.66, p = 3.71 × 10⁻⁹).
Conclusions: We identified DNA methylation signals from multiple tissue types in ALS cfDNA, highlighting diverse tissue involvement in ALS pathology. These findings promote epigenetic cfDNA analysis as a powerful tool for advancing our understanding of neurodegenerative disease.
Keywords: Cell-free DNA; Epigenetics; Neurodegeneration.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All participants provided written informed consent and the study received approval from the Human Research Ethics Committee at the Royal Brisbane and Women’s Hospital (HREC/17/QRBW/299) and by the UCSF Committee on Human Research (IRB 10–05027). Research conformed to the Declarations of Helsinki. Consent for publication: Written informed consent was obtained from individuals to publish their clinical details. Competing interests: C.C., N.Z., M.M., M.P., and F.C.G. are co-inventors on an international patent application (PCT/US2024/033056, filed June 7, 2024) related to cell-free DNA biomarkers for disease diagnosis and prognosis. N.Z. serves as a scientific advisor and consultant for Dinamo Biotechnologies. The remaining authors declare that they have no competing interests.
Figures




References
-
- Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F, et al. The Origin and Mechanism of Circulating DNA. Ann N Y Acad Sci. 2000;906:161–8. 10.1111/j.1749-6632.2000.tb06608.x. - PubMed
-
- Zhang J, Wu Y, Chen S, Luo Q, Xi H, Li J, et al. Prospective prenatal cell-free DNA screening for genetic conditions of heterogenous etiologies. Nat Med. 2024. 10.1038/s41591-023-02774-x. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

