Comprehensive Virome Analysis of Commercial Lilies in South Korea by RT-PCR, High-Throughput Sequencing, and Phylogenetic Analyses
- PMID: 41096872
- PMCID: PMC12525212
- DOI: 10.3390/ijms26199598
Comprehensive Virome Analysis of Commercial Lilies in South Korea by RT-PCR, High-Throughput Sequencing, and Phylogenetic Analyses
Abstract
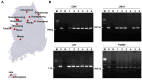
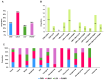
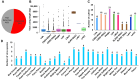
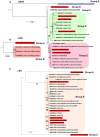
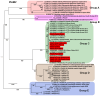
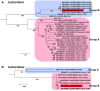
Viral diseases pose a significant threat to lily (Lilium spp.) cultivation; however, large-scale assessments of virus prevalence and diversity in South Korea are limited. This study combined RT-PCR surveys, high-throughput sequencing (HTS), and analyses of 48 lily hybrid transcriptomes to characterize the lily virome. RT-PCR screening of 100 samples from 13 regions showed that 87% were infected, primarily with lily mottle virus (LMoV, 65%), Plantago asiatica mosaic virus (PlAMV, 34%), cucumber mosaic virus (CMV, 34%), and lily symptomless virus (LSV, 25%). Mixed infections were approximately twice as frequent as single infections and were associated with greater symptom severity, particularly in triple-virus combinations. High-throughput sequencing expanded detection to six viruses, including milk vetch dwarf virus (MDV) and lily virus B (LVB), the latter confirmed as a variant of strawberry latent ringspot virus (SLRSV). Near-complete genomes of several viruses were assembled and validated through RT-PCR. Transcriptome mining identified eight virus species across 26 cultivars; PlAMV was the most common, and viral loads varied significantly among hybrids. Phylogenetic analyses revealed close relationships between Korean and Chinese isolates and host-related clustering in PlAMV. These findings highlight the complexity of lily viromes in South Korea and provide essential resources for diagnostics, disease management, and biosecurity.
Keywords: RT-PCR; high-throughput sequencing; lilies; plant viruses; transcriptome analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Kim W.H., Park P.H., Jung J.A., Park K.Y., Suh J.-N., Kwon O.K., Yoo B.S., Lee S.Y., Park P.M., Choi Y.J. Achievement of flower breeding in Korea and its prospects. Korean Soc. Breed. Sci. 2020;52:161–169. doi: 10.9787/KJBS.2020.52.S.161. - DOI
-
- Yang W., Wang P., Zhang W., Xu M., Yan L., Yan Z., Du W., Ouyang L., Liu B., Wu Z. Review on preservation techniques of edible lily bulbs in China. CyTA-J. Food. 2022;20:172–182. doi: 10.1080/19476337.2022.2107708. - DOI
-
- Yasemin S., Beruto M. A review on flower bulb micropropagation: Challenges and opportunities. Horticulturae. 2024;10:284. doi: 10.3390/horticulturae10030284. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

