Drug-disease networks and drug repurposing
- PMID: 41100566
- PMCID: PMC12548869
- DOI: 10.1371/journal.pcbi.1013595
Drug-disease networks and drug repurposing
Abstract
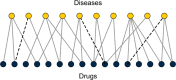
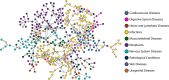
Repurposing existing drugs to treat new diseases is a cost-effective alternative to de novo drug development, but there are millions of potential drug-disease combinations to be considered with only a small fraction being viable. In silico predictions of drug-disease associations can be invaluable for reducing the size of the search space. In this work we present a novel network of drugs and the diseases they treat, compiled using a combination of existing textual and machine-readable databases, natural-language processing tools, and hand curation, and analyze it using network-based link prediction methods to identify potential drug-disease combinations. We measure the efficacy of these methods using cross-validation tests and find that several methods, particularly those based on graph embedding and network model fitting, achieve impressive prediction performance, significantly better than previous approaches, with area under the ROC curve above 0.95 and average precision almost a thousand times better than chance.
Copyright: © 2025 Polanco, Newman. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources

