Loss of O-GlcNAcylation in cardiac myocytes triggers the integrated stress response, contributing to heart failure
- PMID: 41101503
- PMCID: PMC12661449
- DOI: 10.1016/j.jbc.2025.110818
Loss of O-GlcNAcylation in cardiac myocytes triggers the integrated stress response, contributing to heart failure
Abstract
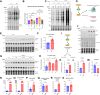
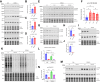
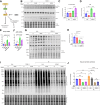
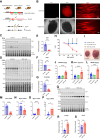
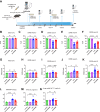
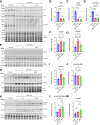
Heart failure (HF) is a significant global health problem, affecting an estimated 64 million people worldwide. At the core of HF is the progressive dysfunction and irreversible loss of cardiac myocytes. O-GlcNAc transferase (OGT) is a conserved enzyme that catalyzes the addition of N-acetyl-glucosamine (GlcNAc) to serine or threonine residues of intracellular proteins. This dynamic protein modification, termed O-GlcNAcylation, has been implicated in nutrient sensing, metabolic regulation and stress adaptation. The integrated stress response (ISR) is a pathway that enables cells to rapidly respond to acute environmental changes and cell damage. During ISR, the translation factor eIF2α is phosphorylated, shutting down general translation but favoring the rapid production of stress-adaptive proteins. However, prolonged activation of the ISR can be detrimental to cells. In this study, we found that inhibiting OGT activates the GCN2/eIF2α/Atf4 signaling axis of the ISR. Activation of this pathway could be blocked by ISRIB, a small molecule that opposes the activity of phosphorylated eIF2α. Mice with inducible deletion of OGT in adult cardiomyocytes developed HF, and treatment with ISRIB significantly delayed the progression to HF. Our study reveals the regulatory impact of O-GlcNAcylation on the ISR and highlights a new potential strategy for alleviating HF.
Keywords: Atf4; GCN2; ISRIB; OGT; PERK; cardiomyocytes; cardiomyopathy; eIF2α; mTOR; translation initiation.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they do not have any conflicts of interest with the content of this article.
Figures



References
-
- Savarese G., Becher P.M., Lund L.H., Seferovic P., Rosano G.M.C., Coats A.J.S. Global burden of heart failure: a comprehensive and updated review of epidemiology. Cardiovasc. Res. 2023;118:3272–3287. - PubMed
-
- Virani S.S., Alonso A., Aparicio H.J., Benjamin E.J., Bittencourt M.S., Callaway C.W., et al. Heart disease and stroke Statistics-2021 update: a report from the American heart association. Circulation. 2021;143:e254–e743. - PubMed
-
- Levine Z.G., Walker S. The biochemistry of O-GlcNAc transferase: which functions make it essential in mammalian cells? Annu. Rev. Biochem. 2016;85:631–657. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

