Transcriptomic Analyses Unveil Hydrocarbon Degradation Mechanisms in a Novel Polar Rhodococcus sp. Strain R1B_2T From a High Arctic Intertidal Zone Exposed to Ultra-Low Sulphur Fuel Oil
- PMID: 41101783
- PMCID: PMC12530890
- DOI: 10.1111/1758-2229.70218
Transcriptomic Analyses Unveil Hydrocarbon Degradation Mechanisms in a Novel Polar Rhodococcus sp. Strain R1B_2T From a High Arctic Intertidal Zone Exposed to Ultra-Low Sulphur Fuel Oil
Abstract
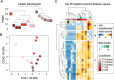
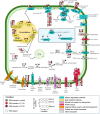
As Arctic shipping increases due to climate change, characterised by rising temperatures and decreasing sea-ice coverage, the risk of oil spills through the Northwest Passage in this fragile ecosystem grows, necessitating effective bioremediation strategies. Research on bioremediation using Arctic coastal sediment bacteria has gained attention, particularly Rhodococcus species that play key roles in hydrocarbon degradation under extreme conditions. This study investigates the hydrocarbon degradation capabilities of a novel cryophilic Arctic Rhodococcus sp. strain R1B_2T isolated from Canadian high Arctic beach sediment in Resolute Bay, exposed to ultra-low sulphur fuel oil for 3 months at 5°C. Comparative transcriptomics analyses revealed dynamic responses and metabolic plasticity, with upregulation of genes for aliphatic, aromatic, and polycyclic aromatic hydrocarbons, biosurfactant production (e.g., rhamnolipid), cold adaptation, and stress responses. The strain possesses several key alkane degradation genes (alkB, almA, CYP153, ladA), with co-expression network analysis highlighting synergistic mechanisms between alkB and CYP153 that target different chain-length alkanes (alkB: ~C5-C20; CYP153: ~C5-C12 and > C30), demonstrating complementary degradation strategies. The findings reveal adaptive mechanisms and degradation kinetics of native Arctic bacteria, highlighting the potential of Arctic cryophilic and halotolerant Rhodococcus species for oil spill remediation in polar marine environments.
Keywords: Rhodococcus; Arctic bacteria; biodegradation; comparative transcriptomic; differential gene expression; hydrocarbon degradation.
© 2025 The Author(s). Environmental Microbiology Reports published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Benjamini, Y. , and Hochberg Y.. 2018. “Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing.” Journal of the Royal Statistical Society, Series B: Statistical Methodology 57: 289–300.
-
- Bento, F. M. , Camargo F. A., Okeke B. C., and Frankenberger W. T.. 2005. “Comparative Bioremediation of Soils Contaminated With Diesel Oil by Natural Attenuation, Biostimulation and Bioaugmentation.” Bioresource Technology 96: 1049–1055. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

