HIFα isoform specific activities drive cell-type specificity of VHL-associated oncogenesis
- PMID: 41102155
- PMCID: PMC12533196
- DOI: 10.1038/s41467-025-64214-3
HIFα isoform specific activities drive cell-type specificity of VHL-associated oncogenesis
Abstract
Cancers arising from dysregulation of generally operative signaling pathways are often tissue specific, but the mechanisms underlying this paradox are poorly understood. Based on striking cell-type specificity, we postulated that these mechanisms must operate early in cancer development and set out to study them in a model of von Hippel Lindau (VHL) disease. Biallelic mutation of the VHL ubiquitin ligase leads to constitutive activation of hypoxia inducible factors HIF1A and HIF2A and is generally a truncal event in clear cell renal carcinoma. We used an oncogenic tagging strategy in which VHL-mutant cells are marked by tdTomato, enabling their observation, retrieval, and analysis early after VHL-inactivation. Here, we reveal markedly different consequences of HIF1A and HIF2A activation, but that both contribute to renal cell-type specific consequences of VHL-inactivation in the kidney. Early involvement of HIF2A in promoting proliferation within the proximal tubular epithelium supports therapeutic targeting of HIF2A early in VHL disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: P.J.R. is a non-executive director of Immunocore Holdings PLC. The remaining authors declare no competing interests.
Figures
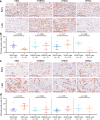

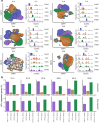

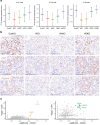
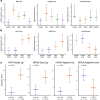
References
-
- Gossage, L., Eisen, T. & Maher, E. R. VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer15, 55–64 (2015). - PubMed
-
- Kaelin, W. G. The von Hippel–Lindau tumor suppressor protein. Annu. Rev. Cancer Biol.2, 91–109 (2018).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

