METTL3-mediated activation of Sonic Hedgehog signaling promotes breast cancer progression
- PMID: 41104016
- PMCID: PMC12521280
- DOI: 10.3389/fcell.2025.1674339
METTL3-mediated activation of Sonic Hedgehog signaling promotes breast cancer progression
Abstract
Introduction: Breast cancer represents a heterogeneous group of tumors characterized by diverse molecular and clinical features, driven by both genetic alterations and epigenetic regulation. Among these mechanisms, the Hedgehog (Hh) developmental pathway, particularly elevated levels of its ligand Sonic Hedgehog (SHH), has been implicated in breast cancer progression. Methyltransferase-like 3 (METTL3), the core catalytic component of the m6A methyltransferase complex, responsible for N6-methyladenosine (m6A) modification of mRNA, has shown a stronger prognostic relevance in regulating mRNA stability and cancer development than other m6A writers, erasers, or readers. Despite evidence suggesting that both SHH and METTL3 contribute to tumor growth in breast tissue, the functional relationship between these factors remains unclear. In this study, we investigated the potential of the METTL3-SHH axis in breast cancer progression to address this gap.
Methods: We have performed bioinformatic analyses by utilizing data from UALCAN, cBioPortal, and GEPIA platforms to comprehensively investigate the methylation patterns, gene expression levels, and mutation profiles of specific genes of interest. Expressions of METTL3 and components of the SHH signaling pathway were analyzed by qRT-PCR. Statistical analyses were performed by using Student's t-test, Spearman and Pearson coefficient (r) test, ANOVA test, and log-rank test.
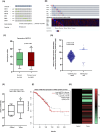
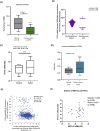
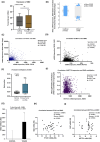
Results: Analysis of 35 breast cancer patients of Bangladesh and gene expression data from The Cancer Genome Atlas (n = 1,021) database revealed METTL3 is overexpressed in breast cancer, and upregulation of METTL3 and downstream key components of the SHH signaling pathway (p < 0.05 vs. control) correlates significantly with worse patient outcomes (HR = 1.3). These findings suggest a possible regulatory mechanism linking METTL3-mediated m6A modification to SHH signaling in breast cancer progression. Elucidating this axis could provide novel insights into tumor biology and identify promising targets for epigenetic therapies.
Keywords: N6-methyladenosine (m6A); breast cancer; epigenetic regulation; methyltransferase like-3 (METTL3); sonic hedgehog developmental pathway.
Copyright © 2025 Baidya, Barua, Shanto, Sonia, Amin, Sultana, Jerin, Jahan, Jahan, Ahmed, Moinul Islam, Parial, Hossain and Noman.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

