PRDM16 regulates smooth muscle cell identity and atherosclerotic plaque composition
- PMID: 41107595
- PMCID: PMC12611775
- DOI: 10.1038/s44161-025-00737-8
PRDM16 regulates smooth muscle cell identity and atherosclerotic plaque composition
Abstract
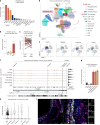
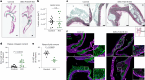
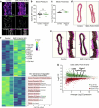
Vascular smooth muscle cells (SMCs) undergo phenotype switching to acquire various fates in response to pathological stimuli. Among these, 'synthetic' SMCs-defined by migration, proliferation and extracellular matrix production-accumulate in atherosclerotic lesions and contribute to fibrous cap formation. The mechanisms driving this synthetic transition remain unclear. Here we identify PRDM16, a gene linked to cardiovascular disease, as a critical transcriptional repressor of the synthetic SMC phenotype. PRDM16 expression declined during SMC modulation, and its deletion in mice induced a synthetic program across all SMC subtypes even without pathological stimuli. Under atherogenic conditions, PRDM16 deficiency resulted in the formation of fibroproliferative plaques with more synthetic SMCs and fewer foam cells. Conversely, enforced PRDM16 expression suppressed SMC migration, proliferation and fibrosis. Mechanistically, PRDM16 occupied chromatin and suppressed activating marks at synthetic loci. These findings establish PRDM16 as a gatekeeper of SMC fate and reveal its role in shaping atherosclerotic plaque composition.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures











Update of
-
PRDM16 controls smooth muscle cell fate in atherosclerosis.bioRxiv [Preprint]. 2025 Feb 23:2025.02.19.639186. doi: 10.1101/2025.02.19.639186. bioRxiv. 2025. Update in: Nat Cardiovasc Res. 2025 Nov;4(11):1573-1588. doi: 10.1038/s44161-025-00737-8. PMID: 40027729 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- UM1 DK126194/DK/NIDDK NIH HHS/United States
- R01HL169766/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- DK123356/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01 DK120982/DK/NIDDK NIH HHS/United States
- R01 HL169766/HL/NHLBI NIH HHS/United States
- R01 HL164577/HL/NHLBI NIH HHS/United States
- R01 HL148239/HL/NHLBI NIH HHS/United States
- DK126194/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01HL148239/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- 40-45200-98-21118/ZonMw (Netherlands Organisation for Health Research and Development)
- R01 HL150359/HL/NHLBI NIH HHS/United States
- DK120982/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01HL150359/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01HL164577/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 DK123356/DK/NIDDK NIH HHS/United States
- R01HL166916/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 HL166916/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
