Stromal cells modulate innate immune cell phenotype and function in colorectal cancer via the Sialic acid/Siglec axis
- PMID: 41111063
- PMCID: PMC12542748
- DOI: 10.1136/jitc-2025-012491
Stromal cells modulate innate immune cell phenotype and function in colorectal cancer via the Sialic acid/Siglec axis
Abstract
Background: The immunosuppressive tumor microenvironment reduces immune response effectiveness in stromal-rich tumors, including consensus molecular subtype 4 colorectal cancer (CRC). Mesenchymal stromal cells (MSCs), precursors to cancer-associated fibroblasts (CAFs), promote cancer progression by suppressing anti-tumor immune responses. Hypersialylation of glycans on tumors engages Siglec receptors on immune cells, driving immune dysfunction, but its role in stromal-mediated suppression of innate immunity remains unclear.
Methods: Sialylation, Sialic acids and Siglec ligands were measured on CRC tissue, primary human normal-associated fibroblasts (NAFs), CAFs, and tumor-conditioned MSCs (MSCTCS) using transcriptional profiles, immunohistochemistry and flow cytometry, respectively. The effect of stromal cell sialylation on macrophages and NK cells was assessed in ex vivo human primary stromal and immune cell co-cultures, and expression of Siglec-10 and immune cell phenotype markers and function was measured by flow cytometry and real-time imaging. Using an immunocompetent Balb/c CT26 mouse model, we induced tumors with/without conditioned stromal cells, with/without pretreatment of stromal cells with sialyltransferase inhibitor (3FAX) or sialidase (E610). We assessed the effect of stromal cell sialylation on macrophages and NK cells in the tumor and secondary lymphoid tissues by flow cytometry.
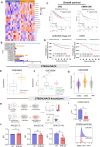
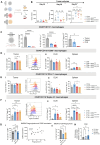
Results: Stromal cells, including CAFs, in CRC tumors are highly sialylated compared with epithelial cancer cells and are associated with high expression of the sialyltransferase ST6GALNAC6. Genetic knockdown of ST6GALNAC6 reduced the expression of stromal cell Siglec-10 ligands in MSCs. CAFs and MSCTCS induced Siglec-10 on macrophages and NK cells and impaired macrophage phagocytosis and NK cell cytotoxicity. Sialidase treatment reduced Siglec-10 expression, restoring macrophage and NK cell antitumor functions. In vivo and ex vivo, desialylation of stromal cells increased macrophage activation (CD11b+CD80+) and reduced immunosuppressive marker expression (CD206, PD-L1, Siglec-G) in lymphoid tissues, indicating sustained systemic anti-tumor immunity. Intratumoral NK cells exhibited high Siglec-G expression and impaired cytotoxicity, and granzyme B expression significantly increased with sialidase treatment of stromal cells. In an inflammatory tumor model, inflammatory tumor-conditioned MSCs (MSCiTCS) promoted metastasis and Siglec-G induction on NK cells and macrophages, both reversed by sialyltransferase inhibition, underscoring the effects of stromal modulation of innate immune cell function in inflammatory tumors.
Conclusions: Stromal cell sialylation modulates innate immune suppression in CRC via the sialic acid/Siglec axis. Targeting stromal sialylation restores NK cytotoxicity and macrophage activation, offering novel insights that may shape therapeutic strategies for reversing immunosuppression in stromal-rich tumors.
Keywords: Colon Cancer; Immune modulatory; Immunosuppression; Sialic acid/Siglec axis; Stroma; Tumor microenvironment - TME.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: LP, JW-YC and LC are employees and shareholders of Palleon Pharmaceuticals. MO’D is a founder of ONK Therapeutics and a member of its Board of Directors and is coinventor on two related patents (US20210186999A1 and US2017032727899A1). TR and AER are coinventors on patent US20210186999A1.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
