Peripheral immune cell-specific genes in Parkinson's disease uncovered by multi-omics with therapeutic implications
- PMID: 41120349
- PMCID: PMC12540756
- DOI: 10.1038/s41531-025-01148-z
Peripheral immune cell-specific genes in Parkinson's disease uncovered by multi-omics with therapeutic implications
Abstract
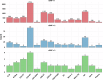
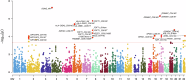
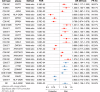
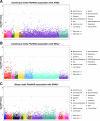
Parkinson's disease (PD) is a complex neurodegenerative disorder with growing evidence suggests peripheral immunity plays a role in its pathogenesis. However, the specific peripheral immune cell types and gene expression profiles associated with PD remain unclear. In this study, we integrated single-cell expression quantitative trait loci (sc-eQTL) data from 14 immune cell types in the OneK1K cohort with large-scale genome-wide association study (GWAS) data for PD. Using Mendelian randomization (MR) and Bayesian colocalization analyses, we identified 28 immune-cell-specific eGenes with significant associations to PD risk, among which 24 showed strong or moderate evidence of shared genetic signals. Notable candidates included FDFT1, ARSA, CTSB, and HLA-DQA1, each displaying cell-type-specific associations in CD4+ T cells, CD8+ T cells, B cells, and monocytes. Replication using an independent sc-eQTL dataset from the DICE project confirmed consistent findings for several eGenes. Additional validation through peripheral blood single-cell RNA sequencing (scRNA-seq) revealed distinct expression patterns and significant changes in PD patients. Phenome-wide association studies (PheWAS) showed multiple associations with immune-related traits and minimal associations with unrelated traits, indicating a favorable safety profile for therapeutic targeting. Drug repurposing analysis identified several candidate compounds, including felodipine, amodiaquine, alprazolam, and tetrandrine, some of which are predicted to cross the blood-brain barrier. Molecular docking simulations further supported strong binding interactions between these compounds and PD-associated targets such as CTSB and ARSA. This integrative approach highlights key immune-cell-specific genes involved in PD and proposes several repurposable drugs with central nervous system potential, paving the way for more targeted therapeutic strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Ascherio, A. & Schwarzschild, M. A. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol.15, 1257–1272 (2016). - PubMed
-
- Poewe, W. et al. Parkinson disease. Nat. Rev. Dis. Prim.3, 17013 (2017). - PubMed
-
- Vijiaratnam, N., Simuni, T., Bandmann, O., Morris, H. R. & Foltynie, T. Progress towards therapies for disease modification in Parkinson’s disease. Lancet Neurol.20, 559–572 (2021). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

