Integrated multi-omics identifies a CD54+ iCAF-ITGAL+ macrophage niche driving immunosuppression via CXCL8-PDL1 axis in cervical cancer
- PMID: 41121145
- PMCID: PMC12539134
- DOI: 10.1186/s12943-025-02471-y
Integrated multi-omics identifies a CD54+ iCAF-ITGAL+ macrophage niche driving immunosuppression via CXCL8-PDL1 axis in cervical cancer
Abstract
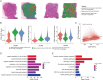
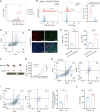
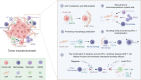
Cervical cancer (CC) remains a formidable clinical challenge, particularly in advanced stages where immune checkpoint blockade yields suboptimal responses. Despite the established role of the tumor microenvironment (TME) in fostering immunosuppression, the precise mechanisms of stroma-immune crosstalk in CC remain elusive. Leveraging single-cell RNA sequencing of 77,221 cells from CC and normal cervical tissues, we uncovered a tumor-enriched subpopulation of inflammatory cancer-associated fibroblasts (iCAFs) marked by elevated CD54 expression (CD54+ iCAFs), which independently predicted adverse clinical outcomes. Systematic dissection of intercellular communication networks revealed a tumor-specific alliance between CD54+ iCAFs and ITGAL+ macrophages, orchestrated through dysregulated ligand-receptor signaling. Spatial multi-omics approaches, including multiplex immunohistochemistry and spatial transcriptomics, confirmed their colocalization within an immunosuppressive niche. Mechanistically, CD54+ iCAFs promote immunosuppression by polarizing ITGAL+ macrophages toward an M2-like phenotype, primarily via CCL2 secretion. These fibroblasts further support immune evasion through two complementary pathways: direct CD54-ITGAL contact-dependent signaling and soluble CCL2-mediated macrophage reprogramming. The resulting macrophage activation stimulates autocrine CXCL8 secretion and subsequent PD-L1 upregulation, which ultimately suppresses CD8+ T cell functions, fostering an immune-tolerant microenvironment in CC. Therapeutic intervention using the CXCL8-CXCR1/2 inhibitor reparixin disrupted the CXCL8-PD-L1 axis, reduced PD-L1+ macrophage abundance and enhanced CD8+ T cell cytotoxicity. Notably, combination therapy with PD-L1 blockade demonstrated synergistic efficacy. Collectively, our findings reveal a stromal-immune checkpoint axis orchestrated by CD54⁺ iCAFs and ITGAL⁺ macrophages that underpins immunosuppression in CC, thereby providing a translational rationale for stroma-directed combination therapies that may overcome resistance to current immunotherapies.
Keywords: CXCL8; Cancer-associated fibroblasts; Cervical cancer; Immunosuppression; Macrophages; PD-L1.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the Research Ethics Committee of the Obstetrics and Gynecology Hospital of Fudan University (Shanghai, China; Ethics Approval No. 2023-49 & 2024-01) and conducted in accordance with the Declaration of Helsinki. All subjects gave written informed consent before participating in the study. Consent for publication: All authors consent to publication. Competing interests: The authors declare no competing interests.
Figures







References
-
- Kalliala I, Athanasiou A, Veroniki AA, Salanti G, Efthimiou O, Raftis N, et al. Incidence and mortality from cervical cancer and other malignancies after treatment of cervical intraepithelial neoplasia: a systematic review and meta-analysis of the literature. Ann Oncol. 2020;31(2):213–27. 10.1016/j.annonc.2019.11.004. - PMC - PubMed
-
- Lorusso D, Xiang Y, Hasegawa K, Scambia G, Leiva M, Ramos-Elias P, et al. Pembrolizumab or placebo with chemoradiotherapy followed by pembrolizumab or placebo for newly diagnosed, high-risk, locally advanced cervical cancer (ENGOT-cx11/GOG-3047/KEYNOTE-A18): a randomised, double-blind, phase 3 clinical trial. Lancet. 2024;403(10434):1341–50. 10.1016/S0140-6736(24)00317-9. - PubMed
-
- O’Malley DM, Neffa M, Monk BJ, Melkadze T, Huang M, Kryzhanivska A, et al. Dual PD-1 and CTLA-4 checkpoint Blockade using balstilimab and Zalifrelimab combination as Second-Line treatment for advanced cervical cancer: an Open-Label phase II study. J Clin Oncol. 2022;740. 10.1200/JCO.21.02067. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 82472993/National Natural Science Foundation of China
- 82173188/National Natural Science Foundation of China
- 21Y11906900/Medical Innovation Research of Shanghai Science and Technology
- 22Y31900500/Medical Innovation Research Special Project under the Science and Technology Innovation Action Plan of the Shanghai Municipal Science and Technology Commission
LinkOut - more resources
Full Text Sources
Medical
Research Materials

