Phenotype switching in highly invasive resistant to vemurafenib and cobimetinib melanoma cells
- PMID: 41121376
- PMCID: PMC12542628
- DOI: 10.1186/s12964-025-02452-0
Phenotype switching in highly invasive resistant to vemurafenib and cobimetinib melanoma cells
Abstract
Background: A mutation in the BRAF (serine/threonine-protein kinase B-raf) gene is most often responsible for the progression of melanoma. A breakthrough in its treatment was the application of BRAF and MEK (mitogen-activated protein kinase kinase) inhibitors. Unfortunately, the effectiveness of this therapy is limited due to rapidly emerging resistance to the drugs. We derived two melanoma cell lines resistant to vemurafenib (a BRAF inhibitor)/cobimetinib (an MEK inhibitor). Due to the significant impact of invasion on cancer progression, we focused our further research on this process.
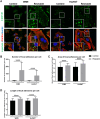
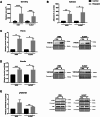
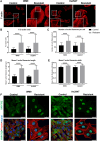
Methods: Cell migration and invasion were assessed via the scratch wound assay. Selected proteins' level as well as the activation of focal adhesion kinase (FAK) were evaluated using Western blotting. The expression of the selected genes was examined by qRT-PCR. The focal adhesions parameters, actin polymerization ratio, as well as YAP/TAZ (Yes-associated protein/transcriptional co-activator with PDZ-binding motif), invadopodia, and β and γ actin localization were analyzed using confocal microscopy. The composition and activity of proteases secreted by cells were determined using a human protease array and gelatin zymography. In addition, cell adhesion and matrix metalloproteinase (MMP14) activity were assessed using appropriate assays.
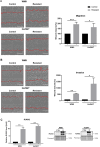
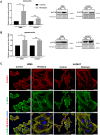
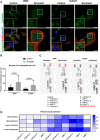
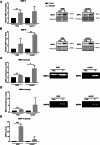
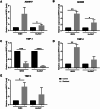
Results: Our analysis showed a greater capacity for migration and invasion of resistant melanoma cells compared to controls, as well as an increase in the level of RUNX2 (runt-related transcription factor 2). Moreover, examined cells exhibited higher adhesion to the surface and were more spread. These cells also formed more focal adhesions. Furthermore, we noticed an increased level of α-parvin and vinculin in resistant cells, as well as an elevated activation of FAK (focal adhesion kinase). Resistance was additionally accompanied by rearrangement of the actin cytoskeleton. Examined cells formed more stress fibers compared to control cells. YAP/TAZ localization became much more nuclear in the resistant ones. The amount of invadopodia was increased, which was reflected by elevated secretion and activation of proteases, as well as altered expression of their inhibitors.
Conclusions: In summary, our study characterized a significantly more invasive phenotype of double-resistant cell lines compared to melanoma cells sensitive to BRAF and MEK inhibitors. Successful inhibition of this phenotype could result in more effective therapy and thus a better prognosis for patients.
Keywords: BRAF inhibitor; Cobimetinib; Drug resistance; MEK inhibitor; Melanoma; Vemurafenib.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
References
-
- Hartman RI, Lin JY. Cutaneous melanoma-a review in detection, staging, and management. Hematol Oncol Clin North Am. 2019;33:25–38. 10.1016/J.HOC.2018.09.005. - PubMed
-
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JWC, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. 10.1038/nature00766. - PubMed
-
- Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, Garbe C, Jouary T, Hauschild A, Grob JJ, Sileni VC, Lebbe C, Mandalà M, Millward M, Arance A, Bondarenko I, a. JB, Haanen G, Hansson J, Utikal J, Ferraresi V, Kovalenko N, Mohr P, Probachai V, Schadendorf D, Nathan P, Robert C, Ribas A, DeMarini DJ, Irani JG, Casey M, Ouellet D, Martin A-M, Le N, Patel K, Flaherty K. Combined BRAF and MEK Inhibition versus BRAF Inhibition alone in Melanoma, N. Engl. J Med. 2014;140929070023009. 10.1056/NEJMoa1406037.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

