Lysosomal damage is a therapeutic target in Duchenne muscular dystrophy
- PMID: 41124255
- PMCID: PMC12542950
- DOI: 10.1126/sciadv.adv6805
Lysosomal damage is a therapeutic target in Duchenne muscular dystrophy
Abstract
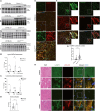
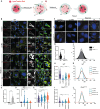
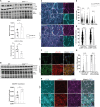
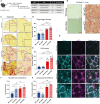
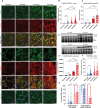
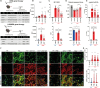
Duchenne muscular dystrophy (DMD), a muscle degenerative disease affecting young boys, arises from the loss of dystrophin. Current gene therapy approaches aim to restore a shortened form of dystrophin (microdystrophin) via adeno-associated vector delivery. While recent clinical studies show promise, therapeutic efficacy remains incomplete, emphasizing the need for improved approaches. Here, we identified lysosomal perturbations in myofibers of patients with DMD and animal models, an overlooked mechanism of cellular damage in muscular dystrophies. These were notably marked by the up-regulation and recruitment of Galectin-3, a biomarker of lysosomal membrane permeabilization, to lysosomes, alongside alterations in lysosome number, morphology, and function. Microdystrophin therapy in Dmdmdx mice fails to fully correct these damages. However, combining it with trehalose, a lysosome-protective disaccharide, substantially improves the outcome, enhancing muscle function, myopathology, and transcriptome. These findings highlight lysosomal damage as an important pathomechanism in DMD and suggest that combining trehalose with gene therapy could enhance therapeutic efficacy.
Figures




References
-
- Roberts T. C., Wood M. J. A., Davies K. E., Therapeutic approaches for Duchenne muscular dystrophy. Nat. Rev. Drug Discov. 22, 917–934 (2023). - PubMed
-
- Bushby K., Finkel R., Birnkrant D. J., Case L. E., Clemens P. R., Cripe L., Kaul A., Kinnett K., McDonald C., Pandya S., Poysky J., Shapiro F., Tomezsko J., Constantin C., DMD Care Considerations Working Group , Diagnosis and management of Duchenne muscular dystrophy, part 1: Diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 9, 77–93 (2010). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

