Bacterial warfare is associated with virulence and antimicrobial resistance
- PMID: 41125589
- PMCID: PMC12546727
- DOI: 10.1038/s41467-025-64363-5
Bacterial warfare is associated with virulence and antimicrobial resistance
Abstract
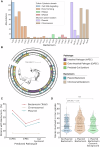
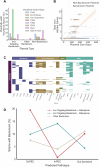
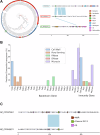
Bacteria have evolved a diverse array of mechanisms to inhibit and kill competitors. However, why some bacteria carry such weapons while others do not remains poorly understood. Here we explore this question using the genomics of the bacteriocins of E. coli as a model system, which have large well-annotated bioinformatic resources. While bacteriocins occur widely, we find that carriage is particularly associated with pathogenic extra-intestinal (ExPEC) strains. These pathogens commonly carry large plasmids encoding bacteriocins alongside virulence factors and antimicrobial resistance mechanisms. Across all strains, we find many orphan immunity proteins, which protect against bacteriocins and suggest that these bacterial weapons are important in nature. We also present evidence that bacteriocin toxins readily move between strains via plasmid transfer and even between plasmids via transposons. Finally, we show that several E. coli bacteriocins are widely shared with the pathogen Salmonella enterica, further cementing the link to virulence. Our work suggests that the bacteriocins of E. coli are important antibacterial weapons for dangerous antimicrobial-resistant strains.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

