The Expression and Clinical Significance of C1orf106 in Low-Grade Serous Ovarian Cancer
- PMID: 41126651
- PMCID: PMC12547486
- DOI: 10.1111/jog.70118
The Expression and Clinical Significance of C1orf106 in Low-Grade Serous Ovarian Cancer
Abstract
Aim: Low-grade serous ovarian cancer (LGSOC) is a rare subtype of ovarian cancer with distinct biological behavior. This study aimed to identify new biomarkers with potential diagnostic and prognostic value for LGSOC.
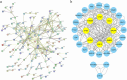
Methods: Gene-expression data were downloaded from the Gene Expression Omnibus (GEO). Differentially expressed genes (DEGs) were identified using R. Functional enrichment analyses were conducted to determine the biological functions and signaling pathways associated with DEGs. The mitogen-activated protein kinase (MAPK) pathway-related gene, chromosome 1 open reading frame 106 (C1orf106), was selected as the target gene. Immunohistochemistry and quantitative real-time polymerase chain reaction (qRT-PCR) were performed to verify its expression. Associations between C1orf106 expression and the clinical features of patients were analyzed using the chi-square (χ2) test. Prognostic significance was evaluated with survival analyses.
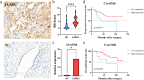
Results: A total of 3099 upregulated and 4968 downregulated genes were identified in LGSOC. Gene set enrichment analysis (GSEA) demonstrated significant alterations in KRAS signaling and metabolic pathways between LGSOC and healthy controls. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses revealed enrichment in immune response and MAPK pathway alterations. Immunohistochemistry and qRT-PCR confirmed that C1orf106 expression in LGSOC tissues was significantly higher than in normal ovarian tissues. Clinically, high C1orf106 expression was associated with lower BMI (< 25 kg/m2), the absence of visible residual disease, and improved progression-free survival (PFS) and overall survival (OS) in univariate Cox and Kaplan-Meier analyses.
Conclusions: C1orf106 may serve as a promising marker for the diagnosis and prognosis of LGSOC.
Keywords: C1orf106; INAVA; bioinformatics; low‐grade serous ovarian cancer; survival analysis.
© 2025 The Author(s). Journal of Obstetrics and Gynaecology Research published by John Wiley & Sons Australia, Ltd on behalf of Japan Society of Obstetrics and Gynecology.
Conflict of interest statement
The authors have nothing to report.
The authors declare no conflicts of interest.
Figures


References
-
- Moujaber T., Ballenine R. L., Gao B., Madsen I., Harnett P. R., and Defazio A., “New Therapeutic Opportunities for Women With Low‐Grade Serous Ovarian Cancer,” Endocrine‐Related Cancer 29, no. 1 (2021): R1–R16. - PubMed
-
- Thomson J. P., Hollis R. L., van Baal J., et al., “Whole Exome Sequencing of Low Grade Serous Ovarian Carcinoma Identifies Genomic Events Associated With Clinical Outcome,” Gynecologic Oncology 174 (2023): 157–166. - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFC2704200/National Key Research and Development Program of China
- 2022YFC2704203/National Key Research and Development Program of China
- 82273348/National Natural Science Foundation of China
- 2024C03159/Science and Technology Program of Zhejiang Province
- 2023AY31021/Science and Technology of the People's Livelihood Project of Jiaxing City
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

