Reanalysis of Undiagnosed Neurodevelopmental Disorder Cases: From RNU4-2 Variants to Clinical Phenotypes
- PMID: 41127311
- PMCID: PMC12539933
- DOI: 10.1212/NXG.0000000000200312
Reanalysis of Undiagnosed Neurodevelopmental Disorder Cases: From RNU4-2 Variants to Clinical Phenotypes
Abstract
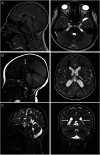
Background and objectives: Neurodevelopmental disorders (NDDs) are often the results of genetic factors, whose identification and key role in their etiology may be elusive. Despite advancements in genetic testing, many cases remain unsolved. Single-nucleotide variants in a critical 18-bp region of the noncoding gene RNU4-2, a crucial component of the spliceosome complex, have been recently recognized as being responsible for ReNU syndrome, also known as NDD with hypotonia, brain anomalies, distinctive facies, and absent language (NEDHAFA). The aim of this study was to investigate the prevalence of RNU4-2 variants within a cohort of unsolved patients exhibiting NDDs from the Telethon Undiagnosed Disease Program (TUDP).
Methods: We conducted a reanalysis of genomic data using bioinformatic tools, followed by direct sequencing to identify variants in the RNU4-2 critical region.
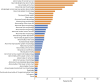
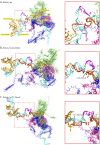
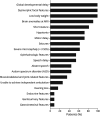
Results: Causative variants were detected in 11 of 375 tested individuals, allowing us to diagnose 2.93% of our unsolved cohort. All heterozygous variants in RNU4-2 occurred de novo, including 10 with the recurrent n.64_65insT insertion and 1 with n.77_78insT. Structural modeling suggested that these variants disrupt the U4/U6 snRNA interaction, potentially impairing spliceosome function.
Discussion: Our findings reinforce the critical role of RNU4-2 variants in syndromic NDDs and underscore the importance of noncoding regions in genetic diagnoses. These findings show that RNU4-2 variants account for approximately 2.9% of the patients with TUDP in this study and highlight the need for integrating advanced molecular techniques and data sharing to refine diagnoses and enhance our understanding of rare genetic disorders.
Copyright © 2025 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology.
Conflict of interest statement
The authors report no relevant disclosures. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
Figures

References
-
- Brown RE. Sex Differences in Neurodevelopment and its Disorders. In: Eisenstat DD, Goldowitz D, Oberlander TF, Yager JY, eds. Neurodev Pediatr. Springer International Publishing; 2023:179-212. Accessed at May 10, 2025. link.springer.com/10.1007/978-3-031-20792-1_11 - DOI
LinkOut - more resources
Full Text Sources
