Bioenergetic reprogramming of macrophages reduces drug tolerance in Mycobacterium tuberculosis
- PMID: 41130947
- PMCID: PMC12549822
- DOI: 10.1038/s41467-025-64407-w
Bioenergetic reprogramming of macrophages reduces drug tolerance in Mycobacterium tuberculosis
Abstract
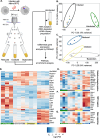
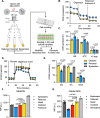
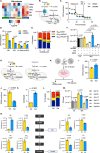
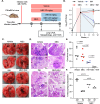
Effective clearance of Mycobacterium tuberculosis (Mtb) requires targeting drug-tolerant populations within host macrophages. Here, we show that macrophage metabolic states govern redox heterogeneity and drug response in intracellular Mtb. Using a redox-sensitive fluorescent reporter (Mrx1-roGFP2), flow cytometry, and transcriptomics, we found that macrophages with high oxidative phosphorylation (OXPHOS) and low glycolysis harbor reductive, drug-tolerant Mtb, whereas glycolytically active macrophages generate mitochondrial ROS via reverse electron transport, imposing oxidative stress on Mtb and enhancing drug efficacy. Computational and genetic analyses identified NRF2 as a key regulator linking host metabolism to bacterial redox state and drug tolerance. Pharmacological reprogramming of macrophages with the FDA-approved drug meclizine (MEC) shifted metabolism towards glycolysis, suppressed redox heterogeneity, and reduced Mtb drug tolerance in macrophages and mice. MEC exhibited no adverse interactions with frontline anti-TB drugs. These findings demonstrate the therapeutic potential of host metabolic reprogramming to overcome Mtb drug tolerance.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

