Reporter-Mediated Evaluation of the Circadian Oscillations of SNAIL Across In Vitro Models
- PMID: 41133664
- PMCID: PMC12550899
- DOI: 10.3390/clockssleep7040054
Reporter-Mediated Evaluation of the Circadian Oscillations of SNAIL Across In Vitro Models
Abstract
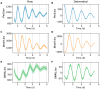
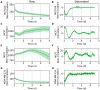
The protein SNAIL has been widely studied for its roles in promoting cancer invasion and resistance to apoptosis. There are multiple contributors to its expression, including self- and circadian regulation, and it has been posited that SNAIL oscillates in a circadian manner. Given the multiple factors involved, we sought to determine whether this is indeed the case. We developed a luciferase reporter that was used to demonstrate SNAIL's rhythmic nature (SNAIL:luc) in the circadian model cell line, U2OS. Considering SNAIL's relevance in breast cancer, we also assessed its oscillations in cellular models representing different levels of aggression. We incorporated the SNAIL:luc reporter in MCF10A breast epithelial cells, and MCF7 and MDA-MB-231 breast cancer cell lines, which are less and more aggressive, respectively. We found that SNAIL oscillations were present but weak in MCF7 and arrhythmic in MDA-MB-231 cells, correlating with those of core clock genes (BMAL1 and PER2) in these models. Surprisingly, MCF10A cells, whose core clock genes possess robust circadian expression patterns, did not have rhythmic oscillations of SNAIL. Our findings suggest that SNAIL is under circadian control, but this is cell line/tissue dependent, setting the stage for additional studies to better understand the impacts of various factors contributing to its expression.
Keywords: MCF10A cells; MCF7 cells; MDA-MB-231 cells; SNAIL; U2OS cells; breast cancer; cancer biology; circadian rhythms; epithelial-to-mesenchymal transition (EMT); luciferase reporters; rhythmicity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

