Mendelian randomization study implicates inflammaging biomarkers in retinal vasculature, cardiovascular diseases, and longevity
- PMID: 41134885
- PMCID: PMC12551708
- DOI: 10.1126/sciadv.adu1985
Mendelian randomization study implicates inflammaging biomarkers in retinal vasculature, cardiovascular diseases, and longevity
Abstract
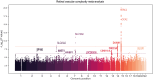
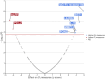
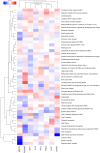
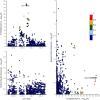
With the increasing proportion of elderly individuals, understanding biological mechanisms of aging is critical. Retinal vascular complexity, measured as fractal dimension (Df) from fundus photographs, has emerged as a vascular aging indicator. We conducted a genome-wide association study of Df on 74,434 participants from the Canadian Longitudinal Study on Aging, Genetics of Diabetes Audit and Research in Tayside Scotland, and UK Biobank cohorts. We identified a novel locus near DAAM1. We found negative genetic correlations between Df and cardiovascular disease, stroke, and inflammation but a positive correlation with life span. By combining the genetic determinants of 1159 circulating proteins from the Prospective Urban and Rural Epidemiological cohort with those of Df using Mendelian randomization, we identified eight causal mediators, including MMP12 and IgG-Fc receptor IIb, which link higher inflammation to lower Df, increased cardiovascular disease risk, and shorter life span. These results extend our understanding of the biological pathways underlying aging processes and inform targets to prevention and treatment.
Conflict of interest statement
H.C.G. holds the McMaster-Sanofi Population Health Institute Chair in Diabetes Research and Care. He reports research grants from Sanofi, Eli Lilly, Novo Nordisk, Abbott, Hanmi, and Boehringer Ingelheim; continuing education grants from Eli Lilly, Abbott, Sanofi, Novo Nordisk, and Boehringer Ingelheim; honoraria for speaking from AstraZeneca, Zuellig, and Jiangsu Hanson; and consulting or advisory board fees from Abbott, Shionogi, Zealand, Pfizer, Novo Nordisk, Eli Lilly, Bayer, and Biolinq, outside the submitted work. E.P.-C started working at the Regeneron Genetics Center LLC during the completion of this study. No data from Regeneron was used in the study. A.V.-V., N.P., Y.H., M.C., E.T., M.R.K.M., W.N., J.P., R.P., S.Y., M.O.B., A.T., K.R., G.P., A.D., and M.P. report no competing interests.
Figures

References
-
- The Lancet Public Health , Ageing: A 21st century public health challenge? Lancet Public Health 2, e297 (2017). - PubMed
-
- L. A. Lipsitz, “Aging as a process of complexity loss,” in Complex Systems Science in Biomedicine, Topics in Biomedical Engineering International Book Series, T. S. Deisboeck, J. Y. Kresh, Eds. (Springer, 2006), pp. 641–654; 10.1007/978-0-387-33532-2_28. - DOI
-
- Zekavat S. M., Raghu V. K., Trinder M., Ye Y., Koyama S., Honigberg M. C., Yu Z., Pampana A., Urbut S., Haidermota S., O’Regan D. P., Zhao H., Ellinor P. T., Segrè A. V., Elze T., Wiggs J. L., Martone J., Adelman R. A., Zebardast N., Del Priore L., Wang J. C., Natarajan P., Deep learning of the retina enables phenome- and genome-wide analyses of the microvasculature. Circulation 145, 134–150 (2022). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

