VaMiAnalyzer: an open source, Python-based application for analysis of 3D in vitro vasculogenic mimicry assays
- PMID: 41136932
- PMCID: PMC12553286
- DOI: 10.1186/s12859-025-06280-4
VaMiAnalyzer: an open source, Python-based application for analysis of 3D in vitro vasculogenic mimicry assays
Abstract
Background: Vasculogenic mimicry (VM) is the phenomenon whereby non-vascular tumor cells develop vascular-like structures. VM is linked to more aggressive tumor phenotypes including higher rates of metastasis and invasion and is potentially resistant to anti-angiogenic cancer therapies. VM is investigated in vitro using 3D assays with microscopy images capturing the resulting VM structures, including loops, branch points, and tubes. The standard method to quantify endpoint data is to count various structural features manually, which is time-consuming and open to bias. At present, no software solutions have been developed to specifically address the analysis and quantification of VM structures.
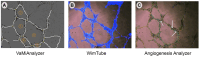
Results: To address this limitation, we developed an open source, Python-based application, VaMiAnalyzer, allowing straightforward quantification of several VM structural features. The application follows a two-step approach that optionally corrects and enhances the raw input images and then analyzes and quantifies the VM features.
Conclusions: VaMiAnalyzer is stand-alone software that allows automated measurement of VM structural features from phase-contrast microscopy images. It produces results that are strongly consistent with manual counts but in a significantly shorter time, allowing quick, non-biased analysis of VM from microscopy images.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Update of
-
VaMiAnalyzer: An open source, python-based application for analysis of 3D in vitro vasculogenic mimicry assays.bioRxiv [Preprint]. 2025 May 14:2025.05.13.653881. doi: 10.1101/2025.05.13.653881. bioRxiv. 2025. Update in: BMC Bioinformatics. 2025 Oct 24;26(1):263. doi: 10.1186/s12859-025-06280-4. PMID: 40463128 Free PMC article. Updated. Preprint.
References
-
- Tonini T, Rossi F, Claudio PP. Molecular basis of angiogenesis and cancer. Oncogene. 2003;22(42):6549–56. - PubMed
-
- Cao Z, Bao M, Miele L, Sarkar FH, Wang Z, Zhou Q. Tumour vasculogenic mimicry is associated with poor prognosis of human cancer patients: a systemic review and meta-analysis. Eur J Cancer. 2013;49(18):3914–23. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

