Estimating (Non)Linear Selection on Reaction Norms: A General Framework for Labile Traits
- PMID: 41140305
- PMCID: PMC12550410
- DOI: 10.1002/ece3.72298
Estimating (Non)Linear Selection on Reaction Norms: A General Framework for Labile Traits
Abstract
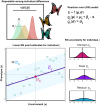
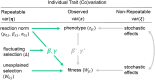
Individual reaction norms describe how labile phenotypes vary as a function of organisms' expected trait values (intercepts) and plasticity across environments (slopes), as well as their degree of stochastic phenotypic variability or predictability (residuals). These reaction norms can be estimated empirically using multilevel, mixed-effects models and play a key role in ecological research on a variety of behavioral, physiological, and morphological traits. Many evolutionary models have also emphasized the importance of understanding reaction norms as a target of selection in heterogeneous and dynamic environments. However, it remains difficult to empirically estimate nonlinear selection on reaction norms, inhibiting robust tests of adaptive theory and accurate predictions of phenotypic evolution. To address this challenge, we propose generalized multilevel models for estimating stabilizing, disruptive, and correlational selection on the reaction norms of labile traits, which can be applied to any repeatedly measured phenotype using a flexible Bayesian framework. Our modeling approach avoids inferential bias by simultaneously accounting for uncertainty in reaction norm parameters and their potentially nonlinear fitness effects. We formally introduce these nonlinear selection models and provide detailed discussion on their interpretation and potential extensions. We then validate their application in a Bayesian framework using simulations. We find that our models facilitate unbiased Bayesian inference across a broad range of effect sizes and desirable power for hypothesis tests with large sample sizes. Coding tutorials are further provided to aid empiricists in applying these models to any phenotype of interest using the Stan probabilistic programming language in R. The proposed modeling framework should, therefore, readily enhance tests of adaptive theory for a variety of labile traits in the wild.
Keywords: complex trait; flexibility; multivariate; personality; phenotypic evolution; plasticity.
© 2025 The Author(s). Ecology and Evolution published by British Ecological Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Acker, P. , Daunt F., Wanless S., et al. 2023. “Hierarchical Variation in Phenotypic Flexibility Across Timescales and Associated Survival Selection Shape the Dynamics of Partial Seasonal Migration.” American Naturalist 201, no. 2: 269–286. - PubMed
-
- Araya‐Ajoy, Y. G. , Dingemanse N. J., Westneat D. F., and Wright J.. 2023. “The Evolutionary Ecology of Variation in Labile Traits: Selection on Its Among‐ and Within‐Individual Components.” Evolution 77: 2246–2256. - PubMed
-
- Araya‐Ajoy, Y. G. , Mathot K. J., and Dingemanse N. J.. 2015. “An Approach to Estimate Short‐Term, Long‐Term and Reaction Norm Repeatability.” Methods in Ecology and Evolution 6, no. 12: 1462–1473.
-
- Araya‐Ajoy, Y. G. , Westneat D. F., and Wright J.. 2020. “Pathways to Social Evolution and Their Evolutionary Feedbacks.” Evolution 74, no. 9: 1894–1907. - PubMed
LinkOut - more resources
Full Text Sources

