Elucidating Mechanisms of Hypomorphic WDR19-Related Kidney Failure
- PMID: 41141533
- PMCID: PMC12545897
- DOI: 10.1016/j.ekir.2025.07.019
Elucidating Mechanisms of Hypomorphic WDR19-Related Kidney Failure
Abstract
Introduction: Variants in the WDR19 gene, a crucial component of the intraflagellar transport (IFT) complex A, are associated with renal-cystic ciliopathies, a prevalent cause of renal failure of genetic origin. In the Arab Druze population, a WDR19 pathogenic missense variant (c.878G>A; p.Cys293Tyr, termed WDR19:C.878G>A) is the most common genetic cause of kidney failure manifesting as adult-onset, typically nonsyndromic chronic kidney disease (CKD). The underlying pathogenesis of this condition remains unclear.
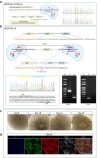
Methods: We used CRISPR-Cas9 to induce patient-specific hypomorphic and loss-of-function (LoF) variants in human embryonic stem cells (hESCs), in addition to using patient-derived induced pluripotent stem cells (iPSCs) for differentiation into kidney organoids. Organoids were assessed by using immunofluorescence, electron microscopy, RNA-sequencing, and pathway analysis to elucidate the effects of these pathogenic variants on kidney development and ciliopathy characteristics.
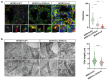
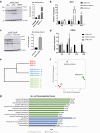
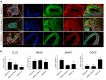
Results: The WDR19 hypomorphic variant impairs nephron development, causing delayed kidney organoid differentiation from early stages, cystogenesis, and structural abnormalities in both tubular and glomerular structures. Mutant organoids displayed reduced ciliation and shortened cilia. Both mutated organoids exhibited Sonic hedgehog dysregulation, where the pathway was upregulated in the presence of severe LoF variant and significantly reduced ciliation. Elevated sonic hedgehog (Shh) signaling was associated with significant downregulation of fibroblast growth factor (FGF) 8 (FGF8) and transcriptomic alterations in associated pathways, suggesting an inverse pathways relationship during kidney organoid development.
Conclusion: Our study validates the pathogenic role of the WDR19 hypomorphic variant in adult-onset renal failure and highlights how hypomorphic pathogenic variants disrupt kidney development. These findings underscore the critical role of cilia in renal development, offering insight into the mechanisms of ciliopathies.
Keywords: WDR19; ciliopathies; kidney organoids.
© 2025 International Society of Nephrology. Published by Elsevier Inc.
Figures


References
LinkOut - more resources
Full Text Sources

