Mass spectrometry-based analysis of rheumatoid factor
- PMID: 41142787
- PMCID: PMC12549570
- DOI: 10.3389/fimmu.2025.1644334
Mass spectrometry-based analysis of rheumatoid factor
Abstract
Introduction: Rheumatoid factor (RF) are autoantibodies that are found in approximately two thirds of patients with rheumatoid arthritis, a chronic autoimmune disease characterized by potentially destructive inflammation of the joints. RF consists of polyclonal antibodies targeting the Fc part of immunoglobulin G. Despite its clinical relevance, RF is not specific for RA, and conventional assays for RF detection, predominantly solid-phase tests detecting IgM RF, suffer from poor harmonization and the disability to test more than one RF isotype.
Methods: We studied RF using a mass-spectrometry-based approach in RF(+), RF(-) rheumatoid arthritis patients and in disease controls. This allowed evaluation of RF at the amino acid level, including the variable and hypervariable region part of RF. RF was captured on Fc coated microwell plates, isolated, digested into peptides and analyzed by liquid chromatography tandem mass spectrometry. An initial proof-of-concept analysis was conducted comprising 12 samples, followed by a larger-scale experiment comprising 86 samples.
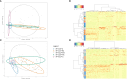
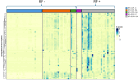
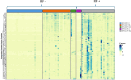
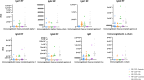
Results: Principal component analysis and sparse partial least squares discriminant analysis demonstrated that RF(+) RA patients displayed peptides that were differentially expressed compared with disease control patients. Framework region-derived peptides, variable region-derived peptides as well as de novo sequenced peptides not present in the human proteome database, were found to be enriched in RF(+) sera compared to disease control sera. Interestingly, some of these peptides were also upregulated in sera from RF(-) RA patients. Furthermore, mass spectrometry analysis revealed different RF isotypes. In addition to IgM, also IgA and IgG isotypes were observed. RF-IgG2 isotype was observed in RF(+) as well as in RF(-) RA patients.
Discussion: In summary, our findings highlight that mass spectrometry provides a platform for elucidating the heterogeneity and isotypic diversity of RF autoantibodies in RA, overcoming limitations inherent to current solid-phase RF assays. Upregulated de novo peptides were found, possibly related to the hypervariable regions of RF. Further validation using integrated proteomic and genomic approaches is required to confirm these novel peptides and their localization within the RF hypervariable regions.
Keywords: complementarity determining regions; immunoglobulins; isotypes; mass spectrometry; rheumatoid factor.
Copyright © 2025 De Leeuw, Michiels, Derua, Dehaemers, Dillaerts, Cockx, Frans, Carpentier, Verschueren and Bossuyt.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

