Moesin controls cell-cell fusion and osteoclast function
- PMID: 41143651
- PMCID: PMC12558046
- DOI: 10.1083/jcb.202409169
Moesin controls cell-cell fusion and osteoclast function
Abstract
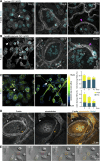
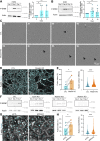
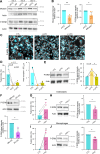
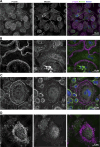
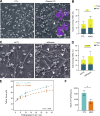
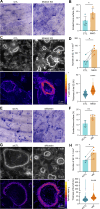
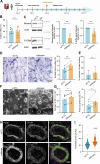
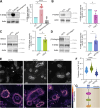
Cell-cell fusion is an evolutionarily conserved process that is essential for many functions, including the formation of bone-resorbing multinucleated osteoclasts. Osteoclast multinucleation involves dynamic interactions between the actin cytoskeleton and the plasma membrane that are still poorly characterized. We found that moesin, a cytoskeletal linker protein member of the Ezrin, radixin, and moesin (ERM) protein family, plays a critical role in both osteoclast fusion and function. Moesin inhibition favors osteoclast multinucleation as well as HIV-1- and inflammation-induced cell fusion. Accordingly, moesin depletion decreases membrane-to-cortex attachment and enhances the formation of tunneling nanotubes, F-actin-based intercellular bridges triggering cell-cell fusion. In addition, moesin regulates the formation of the sealing zone, a key structure determining osteoclast bone resorption area, and thus controls bone degradation via a β3-integrin/RhoA/SLK pathway. Finally, moesin-deficient mice have reduced bone density and increased osteoclast abundance and activity. These findings provide a better understanding of cell-cell fusion and osteoclast biology, opening new opportunities to specifically target osteoclasts in bone disease therapy.
© 2025 Dufrançais et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures





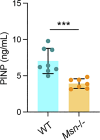
Update of
-
Moesin controls cell-cell fusion and osteoclast function.bioRxiv [Preprint]. 2024 Aug 28:2024.05.13.593799. doi: 10.1101/2024.05.13.593799. bioRxiv. 2024. Update in: J Cell Biol. 2025 Nov 3;224(11):e202409169. doi: 10.1083/jcb.202409169. PMID: 38798563 Free PMC article. Updated. Preprint.
References
-
- Accarias, S., Sanchez T., Labrousse A., Ben-Neji M., Boyance A., Poincloux R., Maridonneau-Parini I., and Le Cabec V.. 2020. Genetic engineering of Hoxb8-immortalized hematopoietic progenitors - A potent tool to study macrophage tissue migration. J. Cell Sci. 133:jcs236703. 10.1242/jcs.236703 - DOI - PubMed
-
- Bagci, H., Sriskandarajah N., Robert A., Boulais J., Elkholi I.E., Tran V., Lin Z.-Y., Thibault M.-P., Dubé N., Faubert D., et al. 2020. Mapping the proximity interaction network of the Rho-family GTPases reveals signalling pathways and regulatory mechanisms. Nat. Cell Biol. 22:120–134. 10.1038/s41556-019-0438-7 - DOI - PubMed
-
- Barrero-Villar, M., Cabrero J.R., Gordón-Alonso M., Barroso-González J., Alvarez-Losada S., Muñoz-Fernández M.A., Sánchez-Madrid F., and Valenzuela-Fernández A.. 2009. Moesin is required for HIV-1-induced CD4-CXCR4 interaction, F-actin redistribution, membrane fusion and viral infection in lymphocytes. J. Cell Sci. 122:103–113. 10.1242/jcs.035873 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- Université Toulouse III - Paul Sabatier (UT3)
- Centre National de la Recherche Scientifique
- 2K12GM081259-16/NH/NIH HHS/United States
- ANR16-CE13-0005-01/French National Research Agency
- APP2029078/National Health & Medical Research Council of Australia
- ANR-18-EURE-0003/French National Research Agency
- ANR-20-CE14-0037/French National Research Agency
- K12 GM081259/GM/NIGMS NIH HHS/United States
- R01 AI147118/AI/NIAID NIH HHS/United States
- EQU202303016313/Fondation pour la Recherche Médicale
- P30 AR069619/AR/NIAMS NIH HHS/United States
- La Ligue contre le Cancer
- P30 AR069619/NH/NIH HHS/United States
- APP2020097/National Health & Medical Research Council of Australia
- Toulouse Université, La Ligue contre le cancer par Toulouse Université, CARe doctoral school
- Institut National de la Santé et de la Recherche Médicale
- Foundation F. Initiativas par Fondation Fonroga Toulouse
- WT_/Wellcome Trust/United Kingdom
- PJT-1620109/CAPMC/ CIHR/Canada
- Fondation Toulouse Cancer Santé
- ANR DFG 2020 JA-3038/2-1/French National Research Agency
- INSERM
- European Molecular Biology Laboratory
- Fondation pour la recherche contre le cancer
- R01 AI147118/NH/NIH HHS/United States
- ANR-10-INBS-04/French National Research Agency
- Toulouse Université Paul Sabatier
LinkOut - more resources
Full Text Sources
Research Materials

