KIAA1429-mediated M6A methylation inhibits osteoclast differentiation via stabilizing Lrp4 mRNA and protects against osteoporosis
- PMID: 41146006
- PMCID: PMC12560588
- DOI: 10.1186/s11658-025-00800-z
KIAA1429-mediated M6A methylation inhibits osteoclast differentiation via stabilizing Lrp4 mRNA and protects against osteoporosis
Abstract
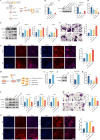
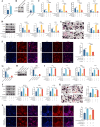
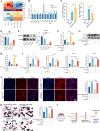
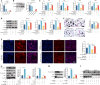
N6-methyladenosine (m6A) is a novel epigenetic modification that has been reported to be involved in the progression of osteoporosis (OP), providing new insights into the pathogenesis of OP. The methyltransferases KIAA1429 [also known as virus-like m6A methyltransferase-associated protein (VIRMA)] participates in various essential biological processes by regulating target gene expression levels. However, the function of KIAA1429-mediated m6A modification in OP progression remains unclear. This study aimed to investigate the biological roles and potential underlying mechanisms of KIAA1429 in OP and osteoclast differentiation. scRNA-seq combined with bulk RNA-seq screening for the differential gene KIAA1429. Analysis of clinical data confirmed KIAA1429 expression and its clinical significance in OP. KIAA1429 inhibited osteoclast differentiation in vitro and reduced bone resorption in ovariectomized (OVX) mice. Mechanistically, LRP4 was identified as a downstream target of KIAA1429. KIAA1429 mediated the m6A modification of Lrp4 mRNA, and then YT521-B homology-domain-containing protein 1 (YTHDC1) increased Lrp4 stability and expression. In addition, LRP4 enhancement recruited TNFAIP3, which inactivated NF-κB signaling. This novel mechanism of NF-κB signaling pathway inhibition by enhanced KIAA1429/YTHDC1-coupled Lrp4 transcription during osteoclast differentiation highlights the potential of KIAA1429 as a novel predictive biomarker and therapeutic target for OP progression.
Keywords: N 6-methyladenosine; KIAA1429; LRP4; Osteoclast; Osteoporosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Human subjects research was approved by the Medical Ethics Committee of Qilu Hospital (Qingdao) of Shandong University (approval no. KYLL-2024036, date: 28 October 2024) and performed in accordance with the principles of the Declaration of Helsinki. Informed consent was obtained from all subjects involved in the study. All animal studies were approved by the Animal Care and Ethics Committee of Shandong University (approval no. 24015, date: 24 April 2024). The Animal Care and Ethics Committee of Shandong University adheres to the principles and guidelines set forth by the International Council for Laboratory Animal Science (ICLAS), ensuring that our animal research meets international ethical standards. Consent for publication: Not applicable. Competing Interests: The authors declare no competing interests.
Figures






References
-
- Ensrud KE, Crandall CJ. Osteoporosis. Ann Intern Med. 2017;167(3):Itc17-itc32. - PubMed
-
- Li J, Chen X, Lu L, Yu X. The relationship between bone marrow adipose tissue and bone metabolism in postmenopausal osteoporosis. Cytokine Growth Factor Rev. 2020;52:88–98. - PubMed
-
- Subarajan P, Arceo-Mendoza RM, Camacho PM. Postmenopausal osteoporosis: a review of latest guidelines. Endocrinol Metab Clin North Am. 2024;53(4):497–512. - PubMed
-
- Compston JE, McClung MR, Leslie WD. Osteoporosis. Lancet. 2019;393(10169):364–76. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

