A luminescence-based biosensor to measure endogenous UBE3A activity
- PMID: 41146712
- PMCID: PMC12554127
- DOI: 10.1016/j.isci.2025.113684
A luminescence-based biosensor to measure endogenous UBE3A activity
Abstract
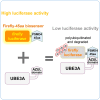
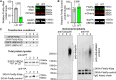
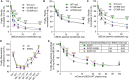
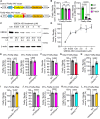
Loss- and gain-of-function (LOF and GOF) mutations in the E3 ubiquitin ligase UBE3A cause distinct neurodevelopmental disorders. The AZUL domain of UBE3A binds with low nanomolar affinity to a 45 amino acid (aa) region of PSMD4. We fused this 45-aa sequence to Firefly luciferase to generate a luminescence-based biosensor, Firefly-45aa, that acts as an artificial UBE3A substrate and exhibits exceptional sensitivity in measuring UBE3A activity. Testing UBE3A variants using Firefly-45aa in HEK293T cells revealed distinct biosensor activity profiles, enabling the classification of LOF and GOF mutations based on maximum activity, inhibitory concentration-50, and activity per unit of protein. Some strong LOF mutations had dominant negative activity. In addition, Firefly-45aa can be used to quantify endogenous UBE3A activity in primary cells from Angelman syndrome and GOF mouse models. This biosensor reveals a mutational spectrum of UBE3A enzyme activity and is a sensitive tool for functional characterization and therapeutic development in UBE3A-related disorders.
Keywords: Cellular neuroscience; Genomics; Neurogenetics.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Weston K.P., Gao X., Zhao J., Kim K.S., Maloney S.E., Gotoff J., Parikh S., Leu Y.C., Wu K.P., Shinawi M., et al. Identification of disease-linked hyperactivating mutations in UBE3A through large-scale functional variant analysis. Nat. Commun. 2021;12:6809. doi: 10.1038/s41467-021-27156-0. - DOI - PMC - PubMed
-
- Xing L., Simon J.M., Ptacek T.S., Yi J.J., Loo L., Mao H., Wolter J.M., McCoy E.S., Paranjape S.R., Taylor-Blake B., Zylka M.J. Autism-linked UBE3A gain-of-function mutation causes interneuron and behavioral phenotypes when inherited maternally or paternally in mice. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112706. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

