Benchmarking cell type and gene set annotation by large language models with AnnDictionary
- PMID: 41152246
- PMCID: PMC12569162
- DOI: 10.1038/s41467-025-64511-x
Benchmarking cell type and gene set annotation by large language models with AnnDictionary
Abstract
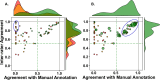
We develop an open-source package called AnnDictionary to facilitate the parallel, independent analysis of multiple anndata. AnnDictionary is built on top of LangChain and AnnData and supports all common large language model (LLM) providers. AnnDictionary only requires 1 line of code to configure or switch the LLM backend and it contains numerous multithreading optimizations to support the analysis of many anndata and large anndata. We use AnnDictionary to perform the first benchmarking study of all major LLMs at de novo cell-type annotation. LLMs vary greatly in absolute agreement with manual annotation based on model size. Inter-LLM agreement also varies with model size. We find that LLM annotation of most major cell types to be more than 80-90% accurate, and will maintain a leaderboard of LLM cell type annotation. Furthermore, we benchmark these LLMs at functional annotation of gene sets, and find that Claude 3.5 Sonnet recovers close matches of functional gene set annotations in over 80% of test sets.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
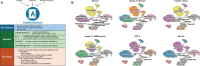



References
-
- Papalexi, E. & Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol.18, 35–45 (2018). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

