Atad1 Is a Potential Candidate Gene for Prepulse Inhibition
- PMID: 41153356
- PMCID: PMC12563959
- DOI: 10.3390/genes16101139
Atad1 Is a Potential Candidate Gene for Prepulse Inhibition
Abstract
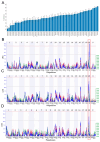
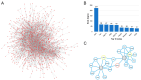
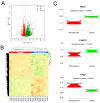
Background/Objectives: Prepulse inhibition (PPI) is a robust, reproducible phenotype associated with schizophrenia and other psychiatric disorders. This study was carried out to identify gene(s) influencing PPI. Methods: We performed Quantitative Trait Locus (QTL) analysis of PPI in 59 strains from the BXD recombinant inbred (BXD RI) mouse family and used a 2-LOD region for candidate gene identification. Genes significantly correlated with the candidate gene were identified based on genetic, partial, and literature correlation, and were further studied through gene enrichment and protein-protein interaction analyses. Phenome-wide association study (PheWAS) and differential expression analyses of the candidate gene were performed using human data. Results: We identified one significant (GN Trait 11428) and two suggestive male-specific QTLs (GN Traits 11426 and 11427) on Chromosome 19 between 27 and 36 Mb with peak LRS values of 19.2 (-logP = 4.2), 14.4 (-logP = 3.1), and 13.3 (-logP = 2.9), respectively. Atad1, ATPase family, AAA domain containing 1 was identified as the strongest candidate for the male-specific PPI loci. Atad1 expression in BXDs is strongly cis-modulated in the nucleus accumbens (NAc, LRS = 26.5 (-logP = 5.7). Many of the Atad1-correlated genes in the NAc were enriched in neurotransmission-related categories. Protein-protein interaction analysis suggested that ATAD1 functions through its direct partners, GRIA2 and ASNA1. PheWAS revealed significant associations between Atad1 and psychiatric traits, including schizophrenia. Analysis of a human RNA-seq dataset revealed differential expression of Atad1 between schizophrenia patients and the control group. Conclusions: Collectively, our analyses support Atad1 as a potential candidate gene for PPI and suggest that this gene should be further investigated for its involvement in psychiatric disorders.
Keywords: Atad1; BXD RI strains; candidate gene; prepulse inhibition; schizophrenia.
Conflict of interest statement
Drs. Bajpai, Freels, Lu, and Cook reported no biomedical financial interests or potential conflicts of interest.
Figures



References
-
- Regier D., Narrow W.E., Rae D.S., Manderscheid R.W., Locke B.Z., Goodwin F.K. The de facto mental and addictive disorders service system. Epidemiologic catchment area prospective 1-year prevalence rates of disorders and services. Arch. Gen. Psychiatry. 1993;50:85–94. doi: 10.1001/archpsyc.1993.01820140007001. - DOI - PubMed
-
- American Psychiatric Association What Is Schizophrenia. 2018. [(accessed on 13 April 2018)]. Available online: https://www.psychiatry.org/patients-families/schizophrenia/what-is-schiz....
-
- Crowley J.J., Sakamoto K. Psychiatric Genomics: Outlook for 2015 and challenges for 2020. Curr. Opin. Behav. Sci. 2015;2:102–107. doi: 10.1016/j.cobeha.2014.12.005. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

