Integrative Methylome and Transcriptome Analysis Reveals Epigenetic Regulation of Pigmentation in Oujiang Color Common Carp
- PMID: 41155296
- PMCID: PMC12562534
- DOI: 10.3390/ijms262010001
Integrative Methylome and Transcriptome Analysis Reveals Epigenetic Regulation of Pigmentation in Oujiang Color Common Carp
Abstract
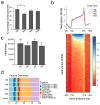
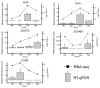
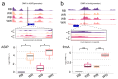
Oujiang color common carp display four striking varieties of pigmentation, but their epigenetic basis is unclear. We integrated genome-wide DNA methylation (MBD-seq) and transcriptomes (RNA-seq) from dorsal skin of four Oujiang color common carp varieties with three biological replicates. Black-spotted groups (RB, WB) showed approximately 6% higher global methylation than non-black-spotted groups (WR, WW), with differential methylation enriched in introns (>23%) and intergenic regions (>47%). Integrative analyses revealed a strong inverse association between promoter methylation and gene expression; 96 pigmentation-related genes were identified, spotlighting genes such as ASIP and frmA as key epigenetically silenced regulators in black-spotted carp. RT-qPCR confirmed directional concordance with RNA-seq for ASIP, frmA, DGAT2, SCARB1, and FOSB. Pathway enrichment implicated melanogenesis metabolism, tyrosine metabolism, lipid metabolism, and fatty acid metabolism, suggesting an interplay between pigment deposition and metabolic regulation. Collectively, the findings present an exploratory view of epigenetic control of coloration and underscore promoter methylation as a core layer influencing color diversity in Oujiang color common carp.
Keywords: MBD-seq; epigenetic regulation; methylation; pigmentation; spot formation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Luo M., Lu G., Yin H., Wang L., Atuganile M., Dong Z. Fish pigmentation and coloration: Molecular mechanisms and aquaculture perspectives. Rev. Aquac. 2021;13:2395–2412. doi: 10.1111/raq.12583. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

