MUC1 promoter methylation pattern diversity and its association with TET3 expression and prognosis in cholangiocarcinoma
- PMID: 41162621
- PMCID: PMC12572258
- DOI: 10.1038/s41598-025-21715-x
MUC1 promoter methylation pattern diversity and its association with TET3 expression and prognosis in cholangiocarcinoma
Abstract
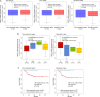
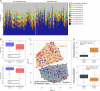
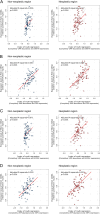
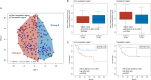
Cholangiocarcinoma (CC) is a highly lethal malignancy that urgently requires reliable prognostic biomarkers. Although MUC1 expression and promoter methylation have been implicated in CC, the clinical significance of promoter methylation pattern composition, beyond average methylation levels, remains unclear. Here, we investigated the relationship between MUC1 promoter methylation heterogeneity, MUC1 mRNA expression, and prognosis in CC. We analyzed bisulfite amplicon sequencing data and mRNA expression of MUC1, DNA methylation-related enzymes (TET1, TET2, TET3, Dnmt1, and Dnmt3a), and tumor microenvironment stress markers in 131 CC tissues. In the neoplastic region, high MUC1 mRNA expression was associated with poor overall survival (HR = 0.131, 95% CI: 0.02 to 0.95, p = 0.042) and correlated with the abundance of completely unmethylated promoter patterns (r = 0.386, p < 0.001). Among the enzymes analyzed, only TET3 expression significantly correlated with the abundance of completely unmethylated patterns in the neoplastic region (Cohen's f2 = 0.108, p = 0.009), suggesting a potential region-specific regulatory association. We visualized beta-diversity in methylation pattern composition using t-SNE and classified samples into two groups based on a linear decision boundary in the t-SNE space. This classification stratified prognosis independently of clinical factors (HR = 0.291, 95% CI: 0.06 to 0.94, p = 0.037; multivariate p = 0.021). These findings propose a novel, composition-based epigenetic stratification framework in CC, revealing that MUC1 promoter methylation pattern structure-rather than average methylation level-has prognostic relevance. Our results highlight the potential of pattern-resolved methylation profiling in the development of clinically applicable epigenetic biomarkers.
Keywords: Cholangiocarcinoma; Diversity; Mucin; Prognosis; Promoter methylation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval and consent to participate: This study was conducted in accordance with the principles of the Declaration of Helsinki. The ethics committee of Kagoshima University Hospital (revised 26–145) approved the IRB application titled “Establishment of a System for the Early Diagnosis and Staging of Pancreatobiliary Cancers Using Pancreatic Juice, Bile, and Duodenal Juice,” which covers the collection, storage, and research use of the tissue samples. Written informed consent was obtained from each patient.
Figures
References
-
- Popescu, I. & Dumitrascu, T. Curative-intent surgery for hilar cholangiocarcinoma: prognostic factors for clinical decision making. Langenbecks Arch. Surg.399, 693–705. 10.1007/s00423-014-1210-x (2014). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

