DNGR-1 signalling limits dendritic cell activation for optimal antigen cross-presentation
- PMID: 41162754
- PMCID: PMC12669754
- DOI: 10.1038/s44318-025-00620-z
DNGR-1 signalling limits dendritic cell activation for optimal antigen cross-presentation
Abstract
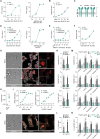
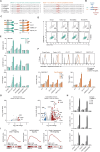
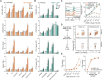
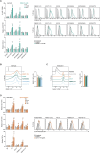
Innate immune receptors often induce activation of conventional dendritic cells (cDCs) and enhance antigen (cross-)presentation, favouring immune responses. DNGR-1 (CLEC9A), a receptor expressed by type 1 cDCs (cDC1s) and implicated in immune responses to viruses and cancer, recognises F-actin exposed on dead cell remnants and promotes cross-presentation of associated antigens. Here, we show that recruitment of phosphatase SHIP1, a process governed by a single amino acid residue adjacent to the signalling motif of the receptor, partly explains how DNGR-1 fails to trigger cDC1 activation in vitro. Substituting this residue converts DNGR-1 into an activating receptor but decreases induction of cross-presentation of dead cell-associated antigens. Introducing the reverse mutation into the related receptor Dectin-1 impairs its activation capacity while enhancing its ability to promote cross-presentation. These findings reveal a functional trade-off in receptor signalling and suggest that DNGR-1 has evolved to prioritise antigen cross-presentation over cellular activation, possibly to minimise inflammatory responses to dead cells.
Keywords: Activation; CLEC9A; Cross-presentation; DNGR-1; cDC1.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. CRS is a founder of Adendra Therapeutics and owns stock options and/or is a paid consultant for Adendra Therapeutics, Montis Biosciences, and Bicycle Therapeutics, all unrelated to this work. CRS also holds appointments as Visiting Professor at Imperial College London and at King’s College London and as honorary professor at University College London. CRS is also a member of the Advisory Editorial Board of The EMBO Journal. This has no bearing on the editorial consideration of this article for publication. The authors declare no competing interests.
Figures





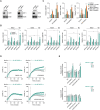

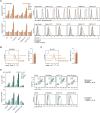
References
-
- Ahrens S, Zelenay S, Sancho D, Hanc P, Kjaer S, Feest C, Fletcher G, Durkin C, Postigo A, Skehel M et al (2012) F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity 36:635–645 - PubMed
-
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001) Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738 - PubMed
-
- Bauer B, Steinle A (2017) HemITAM: a single tyrosine motif that packs a punch. Sci Signal 10:eaan3676 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

