The Microbial Trojan Horse and Antimicrobial Resistance: Acanthamoeba as an Environmental Reservoir for Multidrug Resistant Bacteria
- PMID: 41163086
- PMCID: PMC12572456
- DOI: 10.1111/1462-2920.70193
The Microbial Trojan Horse and Antimicrobial Resistance: Acanthamoeba as an Environmental Reservoir for Multidrug Resistant Bacteria
Abstract
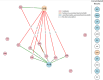
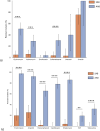
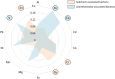
Antimicrobial resistance (AMR) is shaped by environmental pressures, yet the role of microbial predators such as Acanthamoeba in resistance dynamics remains poorly characterized. In this study, Acanthamoeba-associated bacterial communities (AAB) exhibited significantly higher multidrug resistance than sediment-associated bacterial communities (SAB) in a polluted estuarine system. All isolated amoebae belonged to the T4 genotype, suggesting selection for resilient host organisms. AAB displayed elevated multiple antibiotic resistance (MAR) indices and increased resistance to multiple antibiotic classes, particularly aminoglycosides, macrolides, fluoroquinolones and β-lactams. Correlation analysis revealed that resistance in AAB, but not SAB, was associated with potentially toxic elements (PTEs) known to influence phagocyte survival, including arsenic, vanadium, and calcium. These elements may select for traits that confer metal and antibiotic resistance. The findings support a model where protists act as selective environments for AMR, favoring bacteria that possess enhanced tolerance mechanisms. This work provides the first direct evidence linking PTE exposure to the intracellular resistome of Acanthamoeba-associated bacteria. It underscores the need for AMR monitoring frameworks that include protist-bacteria interactions, with implications for One Health and environmental risk assessment strategies. Moreover, this approach is scalable for application in low/middle-income countries, where AMR burden is greatest and surveillance capacity remains limited.
Keywords: Acanthamoeba; Symbiosis; antimicrobial resistance; intracellular; microbiome; potentially toxic elements.
© 2025 The Author(s). Environmental Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Aa, S. , Gnanappazham L., Muraleedharan K. R., et al. 2022. “Multi‐Decadal Changes of Mangrove Forest and Its Response to the Tidal Dynamics of Thane Creek, Mumbai.” Journal of Sea Research 180: 102162. 10.1016/j.seares.2021.102162. - DOI
-
- Ahmad, I. 2022. “Environmental Biofilms as Reservoir of Antibiotic Resistance and Hotspot for Genetic Exchange in Bacteria.” In Beta‐Lactam Resistance in Gram‐Negative Bacteria, 237–265. Springer Nature Singapore.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

