XComposition: multimodal deep learning model to measure body composition using chest radiographs and clinical data
- PMID: 41164313
- PMCID: PMC12560821
- DOI: 10.1093/radadv/umaf035
XComposition: multimodal deep learning model to measure body composition using chest radiographs and clinical data
Abstract
Background: Body composition metrics such as visceral fat volume, subcutaneous fat volume, and skeletal muscle volume are important predictors of cardiovascular disease, diabetes, and cancer prognosis.
Purpose: We explore the use of deep learning to estimate body composition metrics from chest radiographs and a small set of easily obtainable clinical variables.
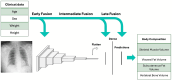
Materials and methods: A retrospective cohort of patients with concurrent noncontrast abdominal CT's and frontal chest radiographs within 3 months of each other was selected. A multitask, multimodal, deep learning model using chest radiographs and clinical variables (age, sex at birth, height and weight extracted from electronic medical records) was trained to estimate the body composition metrics. Reference standard was body composition, including subcutaneous fat volume, measured on CT.
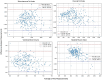
Results: Our final cohort consisted of 1118 patients (582 female and 538 male subjects) from 30 health systems across the United States with imaging performed from 2010 to 2024. The mean age at imaging was 67 years (SD: 17), mean height was 1.67 meters (SD: 0.2), and mean weight was 78 kg (SD: 20). Average values for visceral fat, subcutaneous fat, and skeletal muscle indices were 59.39 cm2/m2 (SD: 39.26), 88.13 cm2/m2 (SD: 58.52), and 44.81 cm2/m2 (SD: 15.49). The best-performing model achieved a Pearson correlation of 0.85 (95% CI: 0.81-0.88) for subcutaneous fat volume, 0.76 (0.65-0.80) for visceral fat volume, and 0.58 (0.49-0.67) for skeletal muscle volume with the multimodal model outperforming unimodal models (P = .0001 for subcutaneous fat volume). Mean absolute errors of the best performing models for subcutaneous and visceral fat volumes were 1054 cm3/m2 and 667 cm3/m2, respectively.
Conclusion: We introduced a multimodal deep learning model leveraging chest radiographs to estimate body composition. Our model can facilitate large-scale studies by estimating body composition using a chest radiograph and commonly available clinical variables.
Keywords: body composition; chest radiographs; deep learning; multimodal data fusion.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Radiological Society of North America.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Iacobini C, Pugliese G, Blasetti Fantauzzi C, Federici M, Menini S. Metabolically healthy versus metabolically unhealthy obesity. Metabolism. 2019;192:51-60. - PubMed
-
- Després JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444(7121):881-887. - PubMed
-
- Santhanam P, Nath T, Peng C, et al. Artificial intelligence and body composition. Diabetes Metab Syndr. 2023;17(3):102732. - PubMed
-
- Pathogenic potential of adipose tissue and metabolic consequences of adipocyte hypertrophy and increased visceral adiposity [Internet]. [cited 2024. Dec 6]. Available from: https://www.tandfonline.com/doi/full/10.1586/14779072.6.3.343 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
