Identification of BET inhibitors (BETi) against solitary fibrous tumor (SFT) through high-throughput screening (HTS)
- PMID: 41166819
- PMCID: PMC12603759
- DOI: 10.1016/j.neo.2025.101244
Identification of BET inhibitors (BETi) against solitary fibrous tumor (SFT) through high-throughput screening (HTS)
Abstract
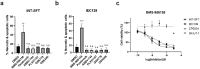
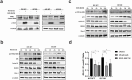
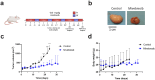
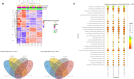
Cancers, especially fusion oncoprotein (FO)-driven hematological cancers and sarcomas, often develop from a low number of key mutations. Solitary Fibrous Tumor (SFT) is a rare mesenchymal tumor driven by the NAB2-STAT6 oncofusion gene. Currently, the treatment options for SFT remain limited, with anti-angiogenic drugs providing only partial responses with an average survival of two years. We constructed SFT cell models harboring specific NAB2-STAT6 fusion transcripts using the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology, and we used these cells as models of SFT. High-throughput drug screens demonstrated that the BET inhibitor Mivebresib can differentially reduce proliferation in SFT cell models. Subsequently, BET inhibitors Mivebresib and BMS-986158 efficiently reduced tumor growth in an SFT patient-derived xenograft (PDX) animal model. Furthermore, our data showed that NAB2-STAT6 fusions may lead to high levels of DNA damage in SFTs. Consequently, combining BET inhibitors with PARP (Poly (ADP-ribose) polymerase) inhibitors or with ATR inhibitors significantly enhanced anti-proliferative effects in SFT cells. Taken together, this study establishes BET inhibitors Mivebresib and BMS-986158 as promising anti-SFT agents.
Keywords: BET inhibitor (BETi); High-throughput screen (HTS); Mivebresib; Solitary fibrous tumor (SFT).
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest David S. Moura has received institutional research grants from PharmaMar and Synox outside the submitted work; travel support from PharmaMar, and personal fees from Tecnopharma, outside the submitted work. Javier Martin-Broto has received honoraria for consulting or advisory board participation and expert testimony from PharmaMar, Bayer, GSK, Deciphera, Boehringer Ingelheim, Cogent Biosciences, Roche, Tecnofarma, and Asofarma; and research funding for clinical studies (institutional) from Deciphera, PharmaMar, Eli Lilly and Company, BMS, Pfizer, Boehringer Ingelheim, Synox, ABBISKO, Biosplice, Lixte, Karyopharm, Rain Therapeutics, INHIBRX, Immunome, Philogen, Cebiotex, PTC Therapeutics, Inc., and SpringWorks Therapeutics. All the other authors do not have competing interests.
Figures



Update of
-
Identification of BET Inhibitors (BETi) Against Solitary Fibrous Tumor (SFT) Through High-Throughput Screening (HTS).bioRxiv [Preprint]. 2025 Mar 28:2025.03.25.645256. doi: 10.1101/2025.03.25.645256. bioRxiv. 2025. Update in: Neoplasia. 2025 Dec;70:101244. doi: 10.1016/j.neo.2025.101244. PMID: 40196499 Free PMC article. Updated. Preprint.
References
-
- Guseva N.V., et al. The NAB2–STAT6 gene fusion in solitary fibrous tumor can be reliably detected by anchored multiplexed PCR for targeted next-generation sequencing. Cancer Genet. 2016;209:303–312. - PubMed
-
- Park Y.S., et al. NAB2-STAT6 fusion protein mediates cell proliferation and oncogenic progression via EGR-1 regulation. Biochem. Biophys. Res. Commun. 2020;526:287–292. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

